Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
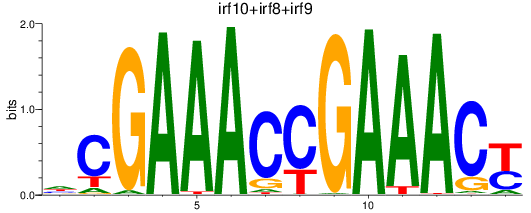
Results for irf10+irf8+irf9
Z-value: 3.93
Transcription factors associated with irf10+irf8+irf9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf9
|
ENSDARG00000016457 | interferon regulatory factor 9 |
|
irf10
|
ENSDARG00000027658 | interferon regulatory factor 10 |
|
irf8
|
ENSDARG00000056407 | interferon regulatory factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf10 | dr11_v1_chr23_+_642395_642395 | 0.86 | 6.1e-02 | Click! |
| irf8 | dr11_v1_chr18_+_30567945_30567945 | 0.58 | 3.0e-01 | Click! |
| irf9 | dr11_v1_chr12_+_13282797_13282807 | 0.50 | 3.9e-01 | Click! |
Activity profile of irf10+irf8+irf9 motif
Sorted Z-values of irf10+irf8+irf9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_13373593 | 4.83 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr11_-_20096018 | 4.71 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr21_-_22648007 | 4.65 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr19_-_7115229 | 4.65 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr1_-_52443379 | 3.56 |
ENSDART00000144676
|
si:ch211-217k17.11
|
si:ch211-217k17.11 |
| chr13_-_37608441 | 3.36 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr19_+_7115223 | 3.34 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr21_-_22673758 | 3.25 |
ENSDART00000164910
|
gig2i
|
grass carp reovirus (GCRV)-induced gene 2i |
| chr21_-_22647695 | 3.22 |
ENSDART00000187553
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr22_-_10580194 | 3.21 |
ENSDART00000105848
|
SHISA4 (1 of many)
|
si:dkey-42i9.7 |
| chr23_+_642395 | 3.19 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr1_-_7951002 | 3.15 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr23_+_642001 | 3.13 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr4_-_76383223 | 3.08 |
ENSDART00000161866
ENSDART00000168456 ENSDART00000174287 ENSDART00000174048 |
zgc:123107
|
zgc:123107 |
| chr22_+_25274712 | 3.08 |
ENSDART00000137341
|
BX950205.1
|
|
| chr22_+_25289360 | 3.07 |
ENSDART00000143782
|
si:ch211-154a22.8
|
si:ch211-154a22.8 |
| chr15_-_17071328 | 3.07 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr3_+_29941777 | 2.96 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr19_-_325584 | 2.94 |
ENSDART00000134266
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr3_+_24603923 | 2.91 |
ENSDART00000172589
|
si:dkey-68o6.6
|
si:dkey-68o6.6 |
| chr1_-_8000428 | 2.85 |
ENSDART00000133098
|
si:dkey-79f11.5
|
si:dkey-79f11.5 |
| chr4_+_7822773 | 2.84 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr25_-_25550938 | 2.76 |
ENSDART00000150412
ENSDART00000103622 |
irf7
|
interferon regulatory factor 7 |
| chr3_+_1942219 | 2.75 |
ENSDART00000114520
|
zgc:165583
|
zgc:165583 |
| chr22_-_17459587 | 2.74 |
ENSDART00000142267
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr10_-_4375190 | 2.70 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr19_-_7110617 | 2.68 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr6_+_52947699 | 2.56 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr22_+_34616151 | 2.53 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr16_+_29514473 | 2.50 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr13_+_22675802 | 2.50 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr4_-_26108053 | 2.49 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr15_-_29162193 | 2.48 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr15_-_21702317 | 2.44 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr13_-_45155792 | 2.39 |
ENSDART00000163556
|
runx3
|
runt-related transcription factor 3 |
| chr13_-_37642890 | 2.34 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr4_-_26095755 | 2.33 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr21_-_22676323 | 2.31 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr4_+_5842433 | 2.23 |
ENSDART00000124085
ENSDART00000179848 |
usp18
|
ubiquitin specific peptidase 18 |
| chr8_-_27656765 | 2.23 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr1_-_25177086 | 2.22 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr5_-_30382925 | 2.21 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr12_-_17028255 | 2.13 |
ENSDART00000152678
|
ifit10
|
interferon-induced protein with tetratricopeptide repeats 10 |
| chr4_-_26107841 | 2.12 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr17_+_35362851 | 2.11 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr16_-_31686602 | 2.09 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr15_+_24588963 | 2.09 |
ENSDART00000155075
|
zgc:198241
|
zgc:198241 |
| chr15_-_47929455 | 2.05 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr6_+_52947186 | 2.03 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr16_+_29509133 | 2.03 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr20_-_16223245 | 2.01 |
ENSDART00000014721
|
echdc2
|
enoyl CoA hydratase domain containing 2 |
| chr7_-_19614916 | 2.01 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr5_+_38662125 | 1.96 |
ENSDART00000136949
|
si:dkey-58f10.13
|
si:dkey-58f10.13 |
| chr11_-_19775182 | 1.92 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr16_-_9802449 | 1.90 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr13_-_37631092 | 1.88 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr9_+_23765587 | 1.88 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr2_-_43135700 | 1.87 |
ENSDART00000098284
|
ftr14
|
finTRIM family, member 14 |
| chr7_-_52417060 | 1.87 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr9_-_23765480 | 1.82 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr16_+_29492937 | 1.82 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr7_+_19615056 | 1.80 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr1_-_21563040 | 1.79 |
ENSDART00000049572
|
ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr1_-_7930679 | 1.72 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr1_-_47161996 | 1.72 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr19_+_23296616 | 1.72 |
ENSDART00000134567
|
irgf1
|
immunity-related GTPase family, f1 |
| chr12_-_23658888 | 1.65 |
ENSDART00000088319
|
map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr15_-_43625549 | 1.63 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr16_+_25285998 | 1.63 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr17_+_45395846 | 1.61 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr25_-_6223567 | 1.60 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr24_+_12945803 | 1.58 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr7_+_3483334 | 1.57 |
ENSDART00000034268
|
si:ch211-285c6.3
|
si:ch211-285c6.3 |
| chr7_+_29509255 | 1.55 |
ENSDART00000076172
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr1_-_8966046 | 1.54 |
ENSDART00000136198
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr22_-_17474583 | 1.54 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr11_+_1584747 | 1.52 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr12_-_30583668 | 1.48 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr4_-_12795030 | 1.45 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr2_+_42708674 | 1.45 |
ENSDART00000141989
ENSDART00000005767 |
ftr11
|
finTRIM family, member 11 |
| chr18_-_38270430 | 1.44 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr4_-_17669881 | 1.44 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr10_+_31809226 | 1.42 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr18_+_26422124 | 1.41 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr3_+_938438 | 1.38 |
ENSDART00000165671
|
irgf4
|
immunity-related GTPase family, f4 |
| chr13_+_27232848 | 1.36 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr22_-_17499513 | 1.34 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr15_+_13984879 | 1.33 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr6_-_49983329 | 1.32 |
ENSDART00000113083
|
asip1
|
agouti signaling protein 1 |
| chr23_-_37536127 | 1.31 |
ENSDART00000078253
|
tor1l1
|
torsin family 1 like 1 |
| chr23_-_4409668 | 1.31 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr25_+_32390794 | 1.31 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr13_-_50200042 | 1.31 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr18_+_13248956 | 1.30 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr5_+_38619813 | 1.28 |
ENSDART00000133314
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr18_-_38270077 | 1.28 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr8_+_47188154 | 1.27 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr3_-_29941357 | 1.27 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr18_-_38270596 | 1.26 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr22_+_19405517 | 1.26 |
ENSDART00000138245
ENSDART00000155144 |
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr5_-_38179220 | 1.25 |
ENSDART00000147701
|
si:ch211-284e13.11
|
si:ch211-284e13.11 |
| chr3_-_1906976 | 1.25 |
ENSDART00000180953
|
zgc:101806
|
zgc:101806 |
| chr22_+_8979955 | 1.25 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr7_+_25053331 | 1.21 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr22_-_6941098 | 1.20 |
ENSDART00000105864
|
zgc:171500
|
zgc:171500 |
| chr5_+_8919698 | 1.18 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr19_+_7124337 | 1.17 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr19_+_4066449 | 1.17 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr9_-_18215644 | 1.17 |
ENSDART00000145876
|
epsti1
|
epithelial stromal interaction 1 |
| chr9_+_11293830 | 1.17 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr3_-_45361573 | 1.17 |
ENSDART00000154796
|
il21r.1
|
interleukin 21 receptor, tandem duplicate 1 |
| chr6_+_59029485 | 1.15 |
ENSDART00000050140
|
CABZ01088367.1
|
|
| chr2_-_37744951 | 1.14 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr17_-_23631400 | 1.11 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr5_-_57723929 | 1.11 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr3_-_32079916 | 1.10 |
ENSDART00000040900
|
baxb
|
BCL2 associated X, apoptosis regulator b |
| chr10_+_17776981 | 1.10 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr5_+_38631119 | 1.06 |
ENSDART00000131832
ENSDART00000162215 |
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr13_+_27232694 | 1.06 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr12_+_38556462 | 1.03 |
ENSDART00000021069
ENSDART00000145377 |
rpl38
|
ribosomal protein L38 |
| chr3_+_6291635 | 1.02 |
ENSDART00000185055
ENSDART00000157707 |
si:ch211-12p12.2
|
si:ch211-12p12.2 |
| chr15_-_5624361 | 1.01 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr2_-_42035250 | 0.99 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr19_+_43878049 | 0.99 |
ENSDART00000165261
|
pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr22_-_8692305 | 0.98 |
ENSDART00000181602
|
CR450686.4
|
|
| chr1_-_44928987 | 0.98 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr3_+_8826905 | 0.95 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr15_+_24572926 | 0.93 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr6_+_58569808 | 0.93 |
ENSDART00000154393
|
helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr16_-_5115993 | 0.92 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr7_+_17908235 | 0.91 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr3_-_7997887 | 0.91 |
ENSDART00000172256
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr23_-_37575030 | 0.90 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr20_+_37393134 | 0.90 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr13_-_50200348 | 0.89 |
ENSDART00000038391
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr22_+_36902352 | 0.88 |
ENSDART00000036273
|
trim35-20
|
tripartite motif containing 35-20 |
| chr9_+_18023288 | 0.87 |
ENSDART00000098355
|
tnfsf11
|
TNF superfamily member 11 |
| chr7_+_3442834 | 0.87 |
ENSDART00000138247
|
si:ch211-285c6.2
|
si:ch211-285c6.2 |
| chr12_-_35936329 | 0.86 |
ENSDART00000166634
|
rnf213b
|
ring finger protein 213b |
| chr1_+_58424507 | 0.86 |
ENSDART00000114197
|
si:ch73-236c18.3
|
si:ch73-236c18.3 |
| chr1_-_10669436 | 0.86 |
ENSDART00000152421
|
si:dkey-31e10.5
|
si:dkey-31e10.5 |
| chr23_-_29357764 | 0.85 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr22_-_17474781 | 0.84 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr1_-_30510839 | 0.84 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr23_-_31266586 | 0.83 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr2_+_41926707 | 0.83 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr8_+_22277198 | 0.81 |
ENSDART00000005989
|
dffb
|
DNA fragmentation factor, beta polypeptide (caspase-activated DNase) |
| chr12_-_17035339 | 0.80 |
ENSDART00000152814
|
ifit9
|
interferon-induced protein with tetratricopeptide repeats 9 |
| chr16_-_24703603 | 0.80 |
ENSDART00000131167
|
negaly6
|
neuromast-expressed gpi-anchored lymphocyte antigen 6 |
| chr13_-_2112450 | 0.80 |
ENSDART00000189343
|
fam83b
|
family with sequence similarity 83, member B |
| chr11_+_25481046 | 0.79 |
ENSDART00000065940
|
opn1lw2
|
opsin 1 (cone pigments), long-wave-sensitive, 2 |
| chr5_+_27421639 | 0.78 |
ENSDART00000146285
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr3_-_22366562 | 0.78 |
ENSDART00000129447
|
ifnphi1
|
interferon phi 1 |
| chr20_-_20402500 | 0.77 |
ENSDART00000190747
ENSDART00000185065 |
prkchb
|
protein kinase C, eta, b |
| chr10_+_26646104 | 0.77 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr12_-_4598237 | 0.75 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chr23_-_31974060 | 0.74 |
ENSDART00000168087
|
BX927210.1
|
|
| chr6_+_32834760 | 0.74 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr6_+_32834007 | 0.73 |
ENSDART00000157353
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr15_+_28355023 | 0.71 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr9_-_35633827 | 0.71 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr23_+_2666944 | 0.70 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr9_+_51147764 | 0.70 |
ENSDART00000188480
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr4_+_68776476 | 0.69 |
ENSDART00000169135
|
si:dkey-264f17.3
|
si:dkey-264f17.3 |
| chr12_-_18578432 | 0.69 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr19_+_4059200 | 0.69 |
ENSDART00000161676
ENSDART00000172424 ENSDART00000161804 |
btr25
|
bloodthirsty-related gene family, member 25 |
| chr2_+_42005475 | 0.69 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr2_-_47904043 | 0.69 |
ENSDART00000185328
ENSDART00000126740 |
ftr22
|
finTRIM family, member 22 |
| chr15_-_28785512 | 0.68 |
ENSDART00000152147
|
blmh
|
bleomycin hydrolase |
| chr25_+_14092871 | 0.68 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr4_-_56673431 | 0.68 |
ENSDART00000161464
ENSDART00000191926 |
si:ch211-227p7.5
|
si:ch211-227p7.5 |
| chr6_-_19271210 | 0.67 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr5_-_67661102 | 0.67 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_+_26645953 | 0.65 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr3_+_36127287 | 0.65 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr14_+_6159356 | 0.65 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr12_-_290413 | 0.65 |
ENSDART00000152496
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr1_+_58332000 | 0.64 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr2_+_16585549 | 0.64 |
ENSDART00000134135
|
si:dkeyp-13a3.10
|
si:dkeyp-13a3.10 |
| chr5_+_28497956 | 0.63 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr24_-_28419444 | 0.63 |
ENSDART00000105749
|
nrros
|
negative regulator of reactive oxygen species |
| chr16_-_43344859 | 0.62 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr15_-_1484795 | 0.61 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr22_-_30678518 | 0.61 |
ENSDART00000138282
|
si:dkey-42l16.1
|
si:dkey-42l16.1 |
| chr22_+_19262161 | 0.61 |
ENSDART00000163654
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr3_-_4455951 | 0.60 |
ENSDART00000193908
ENSDART00000074077 |
trim35-3
|
tripartite motif containing 35-3 |
| chr3_-_7961226 | 0.60 |
ENSDART00000159801
|
trim35-23
|
tripartite motif containing 35-23 |
| chr13_-_2112008 | 0.59 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr16_-_24703855 | 0.59 |
ENSDART00000182422
|
negaly6
|
neuromast-expressed gpi-anchored lymphocyte antigen 6 |
| chr3_+_1724941 | 0.59 |
ENSDART00000193402
|
BX321875.2
|
|
| chr2_+_45479841 | 0.57 |
ENSDART00000151856
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr1_+_58360935 | 0.57 |
ENSDART00000140754
|
si:dkey-222h21.1
|
si:dkey-222h21.1 |
| chr4_-_13567387 | 0.56 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr5_-_45958838 | 0.56 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr19_-_24745317 | 0.55 |
ENSDART00000142774
|
MYO1G
|
si:dkeyp-92c9.3 |
| chr7_-_37563883 | 0.55 |
ENSDART00000148805
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr4_+_35198656 | 0.54 |
ENSDART00000187990
|
si:dkey-269p2.1
|
si:dkey-269p2.1 |
| chr2_-_47870649 | 0.53 |
ENSDART00000142854
|
ftr05
|
finTRIM family, member 5 |
| chr5_-_37820878 | 0.53 |
ENSDART00000165513
|
AL935065.1
|
Danio rerio uncharacterized LOC567472 (LOC567472), mRNA. |
| chr4_+_68939595 | 0.52 |
ENSDART00000168331
ENSDART00000193212 |
si:dkey-264f17.2
|
si:dkey-264f17.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf10+irf8+irf9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.7 | 2.9 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.7 | 2.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.5 | 3.1 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.4 | 1.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 2.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 2.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 1.3 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.4 | 13.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 0.9 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.3 | 0.9 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.3 | 2.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.3 | 1.1 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.2 | 0.7 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 0.6 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.2 | 1.0 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 1.0 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 1.1 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.5 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 1.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 0.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 0.8 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 3.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) antigen processing and presentation of peptide antigen(GO:0048002) |
| 0.2 | 1.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 5.9 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 1.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 1.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 8.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 1.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 0.5 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 1.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.1 | 0.7 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.4 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 1.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 2.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 3.4 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.0 | 4.2 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 2.1 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 16.9 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.1 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 1.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 2.4 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.6 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.1 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.9 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.9 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.8 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.4 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 13.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.4 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 3.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.2 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 9.9 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.8 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 15.1 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.7 | 2.9 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.7 | 2.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.7 | 4.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.5 | 2.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.5 | 10.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 1.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 17.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 2.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.2 | 1.3 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.2 | 1.5 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 5.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 2.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 1.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 12.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.7 | GO:0030250 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 3.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.3 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.8 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.1 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 2.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 7.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.8 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 3.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 5.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 4.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.3 | 1.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 6.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 1.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 1.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 3.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |