Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
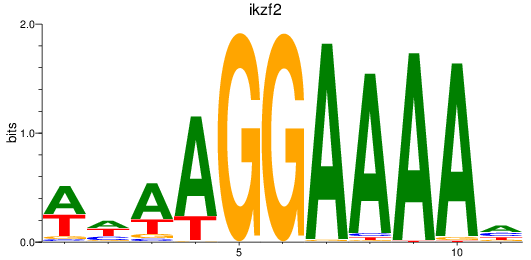
Results for ikzf2
Z-value: 1.27
Transcription factors associated with ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf2
|
ENSDARG00000069111 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | dr11_v1_chr9_-_40014339_40014339 | 0.17 | 7.8e-01 | Click! |
Activity profile of ikzf2 motif
Sorted Z-values of ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_1689775 | 0.88 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 4 |
| chr7_+_39386982 | 0.82 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr20_+_46040666 | 0.82 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr13_+_22264914 | 0.79 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr3_-_61181018 | 0.71 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr16_+_25245857 | 0.68 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr13_-_31647323 | 0.66 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr16_+_23398369 | 0.66 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr16_+_23397785 | 0.65 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr3_+_30921246 | 0.62 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr23_-_32162810 | 0.60 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr9_-_22831836 | 0.57 |
ENSDART00000142585
|
neb
|
nebulin |
| chr3_-_61203203 | 0.55 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr11_+_10548171 | 0.53 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr4_+_7508316 | 0.50 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr5_-_38820046 | 0.50 |
ENSDART00000182886
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr18_+_5543677 | 0.49 |
ENSDART00000146161
ENSDART00000136189 |
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr6_-_46742455 | 0.46 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr21_+_5800306 | 0.46 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr12_+_14084291 | 0.43 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr25_-_18470695 | 0.42 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr8_-_23599096 | 0.41 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr10_-_7785930 | 0.41 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr18_+_5273953 | 0.40 |
ENSDART00000165073
|
slc12a1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr16_+_26774182 | 0.40 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr18_-_16123222 | 0.39 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr2_+_45191049 | 0.39 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr19_-_32804535 | 0.38 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr3_+_23691847 | 0.37 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr23_+_36101185 | 0.37 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr12_-_4070058 | 0.37 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr3_+_14611299 | 0.37 |
ENSDART00000140577
|
tspan35
|
tetraspanin 35 |
| chr24_+_34085940 | 0.36 |
ENSDART00000171189
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr11_+_34235372 | 0.36 |
ENSDART00000063150
|
fam43a
|
family with sequence similarity 43, member A |
| chr23_+_36087219 | 0.35 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr23_+_20408227 | 0.35 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr1_-_59169815 | 0.35 |
ENSDART00000100163
|
wu:fk65c09
|
wu:fk65c09 |
| chr3_-_20091964 | 0.35 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr4_+_76659013 | 0.34 |
ENSDART00000147908
ENSDART00000134229 |
ms4a17a.5
|
membrane-spanning 4-domains, subfamily A, member 17A.5 |
| chr19_-_25519612 | 0.33 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr18_+_26750516 | 0.33 |
ENSDART00000110843
|
alpk3a
|
alpha-kinase 3a |
| chr16_-_31976269 | 0.32 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr15_-_37829160 | 0.32 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr18_+_7591381 | 0.32 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr5_-_68916623 | 0.30 |
ENSDART00000141917
ENSDART00000109053 |
ank1a
|
ankyrin 1, erythrocytic a |
| chr7_-_35432901 | 0.30 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr5_-_68916455 | 0.30 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr5_+_43783360 | 0.29 |
ENSDART00000019431
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr16_-_31756859 | 0.28 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_+_46003449 | 0.28 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr10_+_36662640 | 0.28 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr4_+_18843015 | 0.27 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr1_+_35494837 | 0.27 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr7_+_2455344 | 0.26 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr23_+_45027263 | 0.26 |
ENSDART00000058364
|
hmgb2b
|
high mobility group box 2b |
| chr13_-_22699024 | 0.26 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr24_+_25919809 | 0.25 |
ENSDART00000006615
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr25_-_8602437 | 0.25 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr21_-_11655010 | 0.25 |
ENSDART00000144370
ENSDART00000139814 ENSDART00000139289 |
cast
|
calpastatin |
| chr8_-_21372446 | 0.25 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr25_-_13408760 | 0.24 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr14_-_6666854 | 0.24 |
ENSDART00000133031
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr1_-_55068941 | 0.24 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr13_+_16279890 | 0.24 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr12_-_48477031 | 0.24 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr19_+_46158078 | 0.24 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr22_-_17653143 | 0.24 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr8_-_31062811 | 0.23 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr6_+_39360377 | 0.23 |
ENSDART00000028260
ENSDART00000151322 |
zgc:77517
|
zgc:77517 |
| chr4_+_76775837 | 0.23 |
ENSDART00000174167
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr2_-_24022085 | 0.22 |
ENSDART00000088643
ENSDART00000189822 |
col15a1b
|
collagen, type XV, alpha 1b |
| chr12_+_22576404 | 0.22 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr9_+_5862040 | 0.22 |
ENSDART00000129117
|
pdzk1
|
PDZ domain containing 1 |
| chr16_+_29492749 | 0.22 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr7_+_73822054 | 0.21 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr21_+_25187210 | 0.21 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr1_-_35695614 | 0.21 |
ENSDART00000133813
ENSDART00000145291 ENSDART00000176451 |
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr18_+_19648275 | 0.21 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr5_+_8492640 | 0.21 |
ENSDART00000171701
|
osmr
|
oncostatin M receptor |
| chr3_+_39540014 | 0.20 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr2_+_55984788 | 0.20 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr4_+_25220674 | 0.20 |
ENSDART00000066934
|
itih5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr6_+_49021703 | 0.20 |
ENSDART00000149394
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr20_+_38910214 | 0.20 |
ENSDART00000047362
|
msra
|
methionine sulfoxide reductase A |
| chr7_+_19482084 | 0.20 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr16_+_13427967 | 0.20 |
ENSDART00000038196
|
zgc:101640
|
zgc:101640 |
| chr14_-_46198373 | 0.20 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr11_-_21404358 | 0.19 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr9_+_563547 | 0.19 |
ENSDART00000162761
|
CU984600.2
|
|
| chr20_+_16881883 | 0.19 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr6_-_21534301 | 0.19 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr14_-_14566417 | 0.19 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr17_-_28749640 | 0.19 |
ENSDART00000000948
|
coch
|
coagulation factor C homolog, cochlin (Limulus polyphemus) |
| chr14_-_1280907 | 0.19 |
ENSDART00000186150
|
CABZ01081490.1
|
|
| chr3_+_23692462 | 0.18 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr12_+_38878830 | 0.18 |
ENSDART00000156926
|
si:ch211-39f2.3
|
si:ch211-39f2.3 |
| chr18_-_7481036 | 0.18 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr18_+_26422124 | 0.18 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr13_-_37620091 | 0.18 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr9_+_14010823 | 0.18 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr16_+_25126935 | 0.18 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr19_+_823945 | 0.18 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_+_9966384 | 0.18 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr3_-_34528306 | 0.17 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr15_+_46356879 | 0.17 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr9_+_45839260 | 0.17 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr11_+_20899029 | 0.17 |
ENSDART00000163029
|
zgc:162182
|
zgc:162182 |
| chr20_+_1960092 | 0.17 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr19_+_11217279 | 0.17 |
ENSDART00000181859
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr21_+_5560040 | 0.17 |
ENSDART00000163205
|
si:ch211-134a4.6
|
si:ch211-134a4.6 |
| chr21_+_11248448 | 0.17 |
ENSDART00000142431
|
arid6
|
AT-rich interaction domain 6 |
| chr2_-_45191319 | 0.17 |
ENSDART00000192272
|
CR407590.2
|
|
| chr18_+_44703343 | 0.16 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr13_-_349952 | 0.16 |
ENSDART00000133731
ENSDART00000140326 ENSDART00000189389 ENSDART00000185865 ENSDART00000109634 ENSDART00000147058 ENSDART00000142695 |
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr16_+_38201840 | 0.16 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr1_+_35495368 | 0.16 |
ENSDART00000053806
|
gab1
|
GRB2-associated binding protein 1 |
| chr21_-_13051613 | 0.16 |
ENSDART00000190777
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr12_+_17106117 | 0.16 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr21_+_11385031 | 0.15 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr16_-_26140768 | 0.15 |
ENSDART00000143960
|
cd79a
|
CD79a molecule, immunoglobulin-associated alpha |
| chr6_+_19950107 | 0.15 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr24_-_15263142 | 0.15 |
ENSDART00000183176
ENSDART00000006930 |
rttn
|
rotatin |
| chr17_-_11439815 | 0.15 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr6_-_48347498 | 0.15 |
ENSDART00000149159
|
capza1a
|
capping protein (actin filament) muscle Z-line, alpha 1a |
| chr20_+_26702377 | 0.15 |
ENSDART00000077753
|
foxc1b
|
forkhead box C1b |
| chr25_-_13320986 | 0.15 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr5_+_61361815 | 0.15 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr1_-_52497834 | 0.15 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr14_-_1355544 | 0.15 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr11_-_13126505 | 0.15 |
ENSDART00000158377
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr2_-_37210397 | 0.15 |
ENSDART00000084938
|
apoda.1
|
apolipoprotein Da, duplicate 1 |
| chr3_-_32957702 | 0.15 |
ENSDART00000146586
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr12_+_38929663 | 0.14 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr20_+_38276690 | 0.14 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr12_+_10163585 | 0.14 |
ENSDART00000106191
|
psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr13_-_4223955 | 0.14 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr15_-_951118 | 0.14 |
ENSDART00000171427
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr2_-_31791633 | 0.14 |
ENSDART00000180662
|
retreg1
|
reticulophagy regulator 1 |
| chr16_+_7991274 | 0.14 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr16_-_38333976 | 0.14 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr23_+_36106790 | 0.14 |
ENSDART00000128533
|
hoxc3a
|
homeobox C3a |
| chr10_+_9159279 | 0.14 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr2_-_58183499 | 0.14 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr20_-_44090624 | 0.14 |
ENSDART00000048978
ENSDART00000082283 ENSDART00000082276 |
runx2b
|
runt-related transcription factor 2b |
| chr11_+_18216404 | 0.14 |
ENSDART00000086437
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr12_-_8070969 | 0.14 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr16_+_29492937 | 0.13 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr23_+_24272421 | 0.13 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr14_-_33348221 | 0.13 |
ENSDART00000187749
|
rpl39
|
ribosomal protein L39 |
| chr1_-_1631399 | 0.13 |
ENSDART00000176787
|
clic6
|
chloride intracellular channel 6 |
| chr8_+_47683539 | 0.13 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr16_-_36118514 | 0.13 |
ENSDART00000190427
|
mrps15
|
mitochondrial ribosomal protein S15 |
| chr6_+_54711306 | 0.13 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr7_+_55149001 | 0.13 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr9_-_12659140 | 0.13 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr6_-_40899618 | 0.13 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr14_+_30774515 | 0.13 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr21_-_26495700 | 0.13 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr14_+_33413980 | 0.13 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr19_+_48176745 | 0.13 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr16_-_33095161 | 0.13 |
ENSDART00000187648
|
dopey1
|
dopey family member 1 |
| chr4_-_4250317 | 0.12 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr6_-_23931442 | 0.12 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr6_-_8580857 | 0.12 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr13_-_15994419 | 0.12 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr5_-_28968964 | 0.12 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr5_+_29160324 | 0.12 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr6_-_8489810 | 0.12 |
ENSDART00000124643
|
rasal3
|
RAS protein activator like 3 |
| chr21_+_26657404 | 0.12 |
ENSDART00000129035
ENSDART00000186550 |
prdx5
|
peroxiredoxin 5 |
| chr11_+_43740949 | 0.12 |
ENSDART00000189296
|
CU862021.1
|
|
| chr21_+_31253048 | 0.12 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr9_+_33417969 | 0.12 |
ENSDART00000024795
|
gpr34b
|
G protein-coupled receptor 34b |
| chr21_+_27189490 | 0.12 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr15_-_33834577 | 0.12 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr8_+_47683352 | 0.12 |
ENSDART00000187320
ENSDART00000192605 |
dpp9
|
dipeptidyl-peptidase 9 |
| chr14_+_30753666 | 0.12 |
ENSDART00000009385
ENSDART00000137782 ENSDART00000172778 |
yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr21_-_20329541 | 0.12 |
ENSDART00000166049
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr21_-_21781158 | 0.12 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr21_-_30166097 | 0.12 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr11_-_21404044 | 0.12 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr20_-_26937453 | 0.12 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr16_+_19732543 | 0.12 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr7_-_51368681 | 0.11 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr11_+_29965822 | 0.11 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr4_-_4261673 | 0.11 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr5_-_31942621 | 0.11 |
ENSDART00000049178
|
svopb
|
SV2 related protein b |
| chr20_+_29436601 | 0.11 |
ENSDART00000136804
|
fmn1
|
formin 1 |
| chr7_+_65673885 | 0.11 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr2_-_40191603 | 0.11 |
ENSDART00000180691
|
si:ch211-122l24.6
|
si:ch211-122l24.6 |
| chr8_+_18464235 | 0.11 |
ENSDART00000110571
|
alkal1
|
ALK and LTK ligand 1 |
| chr13_+_24717880 | 0.11 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr14_+_30774894 | 0.11 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr15_-_3963649 | 0.11 |
ENSDART00000172146
ENSDART00000171738 |
gpr171
|
G protein-coupled receptor 171 |
| chr14_+_3522334 | 0.11 |
ENSDART00000164547
|
fnta
|
farnesyltransferase, CAAX box, alpha |
| chr4_+_3980247 | 0.11 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr8_+_10304981 | 0.11 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr11_+_24716837 | 0.11 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr10_-_26179805 | 0.11 |
ENSDART00000174797
|
trim3b
|
tripartite motif containing 3b |
| chr24_+_39990695 | 0.10 |
ENSDART00000040281
|
BX323854.1
|
|
| chr11_-_25418856 | 0.10 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.4 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.6 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.6 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.3 | GO:0030825 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.8 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.3 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.4 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.4 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 1.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.5 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.8 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.0 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.0 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.0 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.0 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |