Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
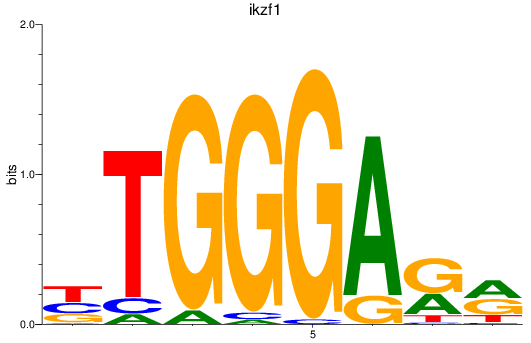
Results for ikzf1
Z-value: 1.43
Transcription factors associated with ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf1
|
ENSDARG00000013539 | IKAROS family zinc finger 1 (Ikaros) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ikzf1 | dr11_v1_chr13_-_15994419_15994472 | 0.27 | 6.6e-01 | Click! |
Activity profile of ikzf1 motif
Sorted Z-values of ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_29161609 | 1.03 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr5_-_71722257 | 0.97 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr5_+_32222303 | 0.77 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr3_-_32817274 | 0.76 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr23_-_23256726 | 0.72 |
ENSDART00000131353
|
si:dkey-98j1.5
|
si:dkey-98j1.5 |
| chr8_-_18535822 | 0.68 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr6_-_39764995 | 0.65 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr13_-_2189761 | 0.63 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr6_-_29195642 | 0.62 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr7_+_39386982 | 0.61 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr25_-_31396479 | 0.58 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr2_-_15324837 | 0.53 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr22_-_282498 | 0.45 |
ENSDART00000182766
|
CABZ01079178.1
|
|
| chr22_-_29191152 | 0.44 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr23_+_6077503 | 0.43 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr22_-_237651 | 0.43 |
ENSDART00000075210
|
zgc:66156
|
zgc:66156 |
| chr7_-_38792543 | 0.43 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr10_-_26163989 | 0.41 |
ENSDART00000136472
|
trim3b
|
tripartite motif containing 3b |
| chr7_-_58098814 | 0.40 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr19_+_22216778 | 0.39 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr13_-_22699024 | 0.38 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr8_+_1009831 | 0.37 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr17_-_14671098 | 0.36 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr21_-_131236 | 0.35 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr2_+_55665322 | 0.35 |
ENSDART00000183636
ENSDART00000183814 |
klf2b
|
Kruppel-like factor 2b |
| chr2_+_55665095 | 0.34 |
ENSDART00000059188
|
klf2b
|
Kruppel-like factor 2b |
| chr9_-_21067971 | 0.34 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr1_-_59116617 | 0.34 |
ENSDART00000137471
ENSDART00000140490 |
MFAP4 (1 of many)
|
si:zfos-2330d3.7 |
| chr7_-_35432901 | 0.34 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr5_-_57641257 | 0.34 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr2_-_21335131 | 0.33 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr17_+_10242166 | 0.33 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr12_+_17100021 | 0.32 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr18_-_16792561 | 0.32 |
ENSDART00000145546
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr17_+_996509 | 0.32 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr2_+_25278107 | 0.31 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr13_-_24825691 | 0.31 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr11_+_10541258 | 0.31 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr13_-_13754091 | 0.31 |
ENSDART00000131255
|
ky
|
kyphoscoliosis peptidase |
| chr11_-_27953135 | 0.30 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr9_-_44642108 | 0.30 |
ENSDART00000086202
|
pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr18_+_20494413 | 0.30 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr7_-_24390879 | 0.30 |
ENSDART00000036680
|
ptgr1
|
prostaglandin reductase 1 |
| chr3_-_59981476 | 0.29 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr3_-_59981162 | 0.28 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr13_-_36391496 | 0.28 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr17_-_43517542 | 0.28 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr15_-_23376541 | 0.27 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr5_+_37517800 | 0.27 |
ENSDART00000048107
|
FP102018.1
|
Danio rerio latent transforming growth factor beta binding protein 3 (ltbp3), mRNA. |
| chr1_-_10914523 | 0.27 |
ENSDART00000007013
|
dmd
|
dystrophin |
| chr4_-_4119396 | 0.27 |
ENSDART00000067409
ENSDART00000138221 |
lmod2b
|
leiomodin 2 (cardiac) b |
| chr7_+_31879649 | 0.27 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr9_-_34260214 | 0.26 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr20_+_20637866 | 0.26 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr25_+_18563476 | 0.26 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr2_-_689047 | 0.26 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr12_+_20641102 | 0.26 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr19_-_42462491 | 0.26 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr17_+_25444323 | 0.25 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr9_-_29985390 | 0.25 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr12_-_26415499 | 0.25 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr24_-_35699595 | 0.25 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_+_12583577 | 0.25 |
ENSDART00000151508
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr24_-_35699444 | 0.24 |
ENSDART00000166567
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_-_26430507 | 0.24 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr7_-_24472991 | 0.24 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr2_+_42724404 | 0.24 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr9_-_21067673 | 0.24 |
ENSDART00000180257
|
tbx15
|
T-box 15 |
| chr21_-_25741096 | 0.23 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr20_+_54738210 | 0.23 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr13_+_24842857 | 0.23 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr1_+_1838164 | 0.23 |
ENSDART00000006013
|
atp1a1a.5
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 5 |
| chr15_-_33933790 | 0.22 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr19_-_40199081 | 0.22 |
ENSDART00000051970
ENSDART00000151079 |
grn2
|
granulin 2 |
| chr13_+_52061034 | 0.22 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr10_-_44008241 | 0.21 |
ENSDART00000137686
|
acads
|
acyl-CoA dehydrogenase short chain |
| chr23_-_26077038 | 0.21 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr18_-_41375120 | 0.21 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr20_+_15982482 | 0.21 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr19_-_38539670 | 0.21 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr21_-_35534401 | 0.21 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr4_-_12914163 | 0.21 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr15_-_33904831 | 0.21 |
ENSDART00000164333
ENSDART00000165404 |
mag
|
myelin associated glycoprotein |
| chr1_+_51496862 | 0.21 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr3_+_41922114 | 0.21 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr19_-_9648542 | 0.20 |
ENSDART00000172628
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr7_+_17816470 | 0.20 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr10_-_8358396 | 0.20 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr7_+_39410180 | 0.20 |
ENSDART00000168641
|
CT030188.1
|
|
| chr1_+_5275811 | 0.20 |
ENSDART00000189676
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr22_+_15898221 | 0.20 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr17_-_33416020 | 0.20 |
ENSDART00000140149
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr17_+_6793001 | 0.20 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr5_-_31712399 | 0.19 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr2_+_58377395 | 0.19 |
ENSDART00000193511
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr12_+_23762966 | 0.19 |
ENSDART00000152942
ENSDART00000181725 |
jcada
|
junctional cadherin 5 associated a |
| chr23_+_34047413 | 0.19 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr17_-_8899323 | 0.19 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr17_+_2549503 | 0.19 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr1_+_26099250 | 0.19 |
ENSDART00000054205
|
ndufb6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 |
| chr13_-_36525982 | 0.19 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr20_+_20638034 | 0.19 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr5_-_31716713 | 0.19 |
ENSDART00000131443
|
dpm2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr15_-_33925851 | 0.18 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr5_+_24287927 | 0.18 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr15_-_47193564 | 0.18 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr17_-_45370200 | 0.18 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr10_-_8129175 | 0.18 |
ENSDART00000133921
|
ndufa9b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9b |
| chr14_-_51855047 | 0.18 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr3_-_7134113 | 0.18 |
ENSDART00000180849
|
BX005085.6
|
|
| chr24_-_28259127 | 0.17 |
ENSDART00000149589
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr20_+_2039518 | 0.17 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr23_+_43668756 | 0.17 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr3_-_52614747 | 0.17 |
ENSDART00000154365
|
trim35-13
|
tripartite motif containing 35-13 |
| chr5_-_26466169 | 0.17 |
ENSDART00000144035
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr2_-_34555945 | 0.17 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr5_+_32817688 | 0.17 |
ENSDART00000139472
|
crata
|
carnitine O-acetyltransferase a |
| chr11_+_6456146 | 0.17 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr23_+_9057999 | 0.16 |
ENSDART00000091899
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr24_-_38816725 | 0.16 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr2_-_32501501 | 0.16 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr20_-_27330383 | 0.16 |
ENSDART00000153277
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr12_-_47782623 | 0.16 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr2_+_35993404 | 0.16 |
ENSDART00000170845
|
lamc2
|
laminin, gamma 2 |
| chr12_+_30109698 | 0.15 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr10_+_9697097 | 0.15 |
ENSDART00000167821
|
rabgap1
|
RAB GTPase activating protein 1 |
| chr23_-_3674443 | 0.15 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr14_+_7048930 | 0.15 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr5_+_19337108 | 0.15 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr14_+_16813816 | 0.15 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr3_+_59411956 | 0.15 |
ENSDART00000166982
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr15_+_15779184 | 0.15 |
ENSDART00000156902
|
si:ch211-33e4.2
|
si:ch211-33e4.2 |
| chr17_+_33415542 | 0.14 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr19_-_7291733 | 0.14 |
ENSDART00000015559
|
sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr10_+_31951338 | 0.14 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr22_-_26834043 | 0.14 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr7_+_39410393 | 0.14 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr19_-_9829965 | 0.14 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr21_+_7582036 | 0.14 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr2_+_3428357 | 0.14 |
ENSDART00000125967
|
CU633991.1
|
|
| chr4_+_2619132 | 0.14 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr3_+_32443395 | 0.14 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr5_-_28606916 | 0.14 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr12_+_23991276 | 0.13 |
ENSDART00000153136
|
psme4b
|
proteasome activator subunit 4b |
| chr22_-_34551568 | 0.13 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr5_-_26181863 | 0.13 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr19_+_24872159 | 0.13 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr22_-_4742866 | 0.13 |
ENSDART00000177992
|
CR751224.1
|
|
| chr2_-_42558549 | 0.13 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr14_-_3268155 | 0.13 |
ENSDART00000177244
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr3_-_58189429 | 0.13 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr7_+_17816006 | 0.13 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr15_-_2640966 | 0.13 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr17_+_24747209 | 0.13 |
ENSDART00000154623
|
stac
|
SH3 and cysteine rich domain |
| chr2_-_14571577 | 0.13 |
ENSDART00000170687
|
pde4bb
|
phosphodiesterase 4B, cAMP-specific b |
| chr8_-_51579286 | 0.13 |
ENSDART00000147878
|
ankrd39
|
ankyrin repeat domain 39 |
| chr17_+_25414033 | 0.13 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr5_+_23598364 | 0.13 |
ENSDART00000132155
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr9_+_33145522 | 0.13 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr17_+_8925232 | 0.13 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr7_+_31130667 | 0.12 |
ENSDART00000173937
|
tjp1a
|
tight junction protein 1a |
| chr9_+_2507526 | 0.12 |
ENSDART00000166579
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr21_+_13182149 | 0.12 |
ENSDART00000140267
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr23_+_21663631 | 0.12 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr7_+_25003313 | 0.12 |
ENSDART00000131935
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr21_-_40835069 | 0.12 |
ENSDART00000004686
|
limk1b
|
LIM domain kinase 1b |
| chr13_-_8229977 | 0.12 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr2_-_32505091 | 0.12 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr7_+_72279584 | 0.12 |
ENSDART00000172021
|
tollip
|
toll interacting protein |
| chr16_-_8927425 | 0.12 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr12_+_17042754 | 0.12 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr16_+_37470717 | 0.12 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr2_-_21847935 | 0.12 |
ENSDART00000003940
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr6_-_54107269 | 0.12 |
ENSDART00000190017
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr24_+_35947077 | 0.12 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr18_+_20869923 | 0.12 |
ENSDART00000138471
|
pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr8_+_30671060 | 0.11 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr3_+_27665160 | 0.11 |
ENSDART00000103660
|
clcn7
|
chloride channel 7 |
| chr2_-_31936966 | 0.11 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr21_+_6290566 | 0.11 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr23_+_2361184 | 0.11 |
ENSDART00000184469
|
CABZ01048666.1
|
|
| chr19_+_33732487 | 0.11 |
ENSDART00000010294
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr13_+_28735151 | 0.11 |
ENSDART00000150376
|
ldb1a
|
LIM domain binding 1a |
| chr3_-_21288202 | 0.11 |
ENSDART00000191766
ENSDART00000187319 |
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr25_-_31763897 | 0.11 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr16_+_1353894 | 0.11 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr10_+_31953502 | 0.11 |
ENSDART00000185634
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr25_-_20238793 | 0.11 |
ENSDART00000145987
|
dnm1l
|
dynamin 1-like |
| chr9_+_55857193 | 0.11 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr15_+_16908085 | 0.11 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr12_-_7824291 | 0.11 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr25_-_23526058 | 0.11 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr1_+_36194761 | 0.11 |
ENSDART00000053773
ENSDART00000147458 |
lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_45076131 | 0.11 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr10_+_21786656 | 0.11 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr23_-_20002459 | 0.11 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr14_+_23158021 | 0.11 |
ENSDART00000084664
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr20_-_26421112 | 0.11 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr13_-_46991577 | 0.11 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr3_+_40170216 | 0.11 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 1.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.2 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.7 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.5 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.2 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0033605 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.2 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.2 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0035176 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0048713 | positive regulation of glial cell differentiation(GO:0045687) regulation of oligodendrocyte differentiation(GO:0048713) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.0 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0044819 | mitotic G1/S transition checkpoint(GO:0044819) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.8 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 1.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 0.3 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.7 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.4 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |