Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
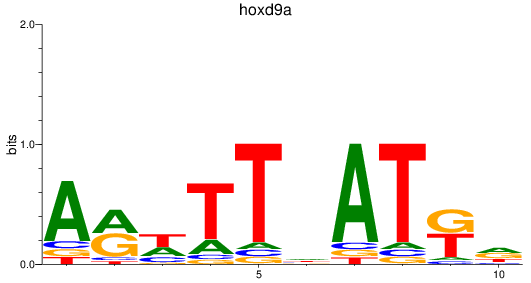
Results for hoxd9a
Z-value: 0.79
Transcription factors associated with hoxd9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd9a
|
ENSDARG00000059274 | homeobox D9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd9a | dr11_v1_chr9_-_1965727_1965727 | 0.02 | 9.7e-01 | Click! |
Activity profile of hoxd9a motif
Sorted Z-values of hoxd9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_46425262 | 0.85 |
ENSDART00000153690
|
sc:d0202
|
sc:d0202 |
| chr6_-_53426773 | 0.80 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr13_+_17672527 | 0.54 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr7_+_56577522 | 0.54 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr14_+_32430982 | 0.51 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr22_+_37888249 | 0.50 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr22_-_24880824 | 0.50 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr25_+_31276842 | 0.49 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr2_+_27855346 | 0.49 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr11_+_37201483 | 0.49 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr3_+_1030479 | 0.46 |
ENSDART00000143449
|
zgc:172253
|
zgc:172253 |
| chr15_+_14856307 | 0.45 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr20_-_43741159 | 0.45 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr19_+_2876106 | 0.44 |
ENSDART00000189309
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr24_-_38083378 | 0.42 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr16_+_23913943 | 0.41 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr2_+_11031360 | 0.39 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr5_+_28830643 | 0.38 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr22_+_12770877 | 0.37 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr6_+_8626427 | 0.37 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr23_+_37482727 | 0.37 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr14_+_29975268 | 0.37 |
ENSDART00000172985
|
cyp4v7
|
cytochrome P450, family 4, subfamily V, polypeptide 7 |
| chr7_-_51953807 | 0.35 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr2_+_27855102 | 0.34 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr19_+_10855158 | 0.32 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr6_-_7776612 | 0.32 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr2_-_39558643 | 0.32 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr23_+_2666944 | 0.32 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr4_+_14899720 | 0.31 |
ENSDART00000187456
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr19_-_657439 | 0.31 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr16_+_1383914 | 0.31 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr15_-_32383340 | 0.31 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr11_-_25213651 | 0.30 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr5_-_1963498 | 0.30 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr10_+_23537576 | 0.30 |
ENSDART00000130515
|
CR847844.1
|
|
| chr20_-_33515890 | 0.29 |
ENSDART00000159987
|
si:dkey-65b13.13
|
si:dkey-65b13.13 |
| chr6_+_41191482 | 0.29 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr17_+_20174812 | 0.29 |
ENSDART00000186628
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr22_+_1049367 | 0.28 |
ENSDART00000065380
|
camk1ga
|
calcium/calmodulin-dependent protein kinase IGa |
| chr20_+_18163821 | 0.26 |
ENSDART00000186507
|
aqp4
|
aquaporin 4 |
| chr22_+_2959879 | 0.26 |
ENSDART00000185354
ENSDART00000063545 |
impact
|
impact RWD domain protein |
| chr17_-_14876758 | 0.25 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr12_+_13164706 | 0.25 |
ENSDART00000112565
|
paqr4b
|
progestin and adipoQ receptor family member IVb |
| chr10_+_14963898 | 0.25 |
ENSDART00000187363
ENSDART00000175732 |
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr21_-_42831033 | 0.24 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr6_-_52675630 | 0.24 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr15_-_32383529 | 0.24 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr22_+_1028724 | 0.24 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr5_-_9216758 | 0.24 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr24_-_25166416 | 0.23 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr25_+_31227747 | 0.23 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr8_+_42718135 | 0.23 |
ENSDART00000158900
|
zgc:194007
|
zgc:194007 |
| chr11_-_4787083 | 0.22 |
ENSDART00000159425
|
FHIT
|
si:ch211-63i20.3 |
| chr10_-_24868581 | 0.22 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr6_-_34938678 | 0.22 |
ENSDART00000186689
ENSDART00000131610 |
serbp1a
|
SERPINE1 mRNA binding protein 1a |
| chr3_+_32135037 | 0.22 |
ENSDART00000110490
|
BX511021.1
|
|
| chr5_-_23805387 | 0.22 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr7_-_16194952 | 0.21 |
ENSDART00000173739
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr6_-_58764672 | 0.21 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr11_+_43431289 | 0.21 |
ENSDART00000192526
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr8_-_40075983 | 0.20 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr17_+_30442518 | 0.20 |
ENSDART00000155264
|
lpin1
|
lipin 1 |
| chr5_-_12093618 | 0.20 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr22_-_16154771 | 0.19 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr25_+_418932 | 0.19 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr3_+_21225750 | 0.19 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr4_+_14900042 | 0.19 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr1_-_40140424 | 0.19 |
ENSDART00000148234
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr12_-_26407092 | 0.19 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr13_-_24260609 | 0.19 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr13_-_39160018 | 0.18 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr24_-_21090447 | 0.18 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr25_+_36328524 | 0.18 |
ENSDART00000073404
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr3_-_49382896 | 0.18 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr13_-_24257631 | 0.18 |
ENSDART00000146524
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr13_-_9070754 | 0.18 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr25_-_4192637 | 0.18 |
ENSDART00000153832
|
ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr10_+_26612321 | 0.18 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr20_-_35076387 | 0.18 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr7_-_24046999 | 0.18 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr12_-_990149 | 0.17 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr1_+_41898919 | 0.17 |
ENSDART00000140845
|
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr21_+_6556635 | 0.17 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr5_+_45007962 | 0.17 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr22_+_9027884 | 0.17 |
ENSDART00000171839
|
si:ch211-213a13.2
|
si:ch211-213a13.2 |
| chr22_-_31059670 | 0.17 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr16_-_16761164 | 0.17 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr17_-_45383699 | 0.16 |
ENSDART00000141182
|
tmem206
|
transmembrane protein 206 |
| chr7_+_38510197 | 0.16 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr15_-_17813680 | 0.16 |
ENSDART00000158556
|
CT573342.2
|
|
| chr24_-_1985007 | 0.16 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr11_+_11303458 | 0.16 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr25_-_30383617 | 0.15 |
ENSDART00000165342
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr13_+_45709380 | 0.15 |
ENSDART00000192862
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr4_+_2267641 | 0.15 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr13_-_37229245 | 0.15 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr9_+_21259820 | 0.15 |
ENSDART00000137024
ENSDART00000132324 |
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr8_-_46455874 | 0.15 |
ENSDART00000146985
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr6_+_43400059 | 0.15 |
ENSDART00000143374
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr15_+_15390882 | 0.15 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr23_+_4483083 | 0.15 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr15_+_17406920 | 0.14 |
ENSDART00000081059
|
rps6kb1b
|
ribosomal protein S6 kinase b, polypeptide 1b |
| chr10_+_40700311 | 0.14 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr12_-_30359498 | 0.14 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr25_+_35019693 | 0.14 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr22_+_16555939 | 0.14 |
ENSDART00000012604
|
pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr1_+_40801696 | 0.14 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr11_-_3366782 | 0.14 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr6_+_59984772 | 0.14 |
ENSDART00000048449
|
mtrf1
|
mitochondrial translational release factor 1 |
| chr15_-_34322915 | 0.14 |
ENSDART00000190543
ENSDART00000019651 ENSDART00000193601 |
dgkb
|
diacylglycerol kinase, beta |
| chr7_-_24047316 | 0.14 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr2_+_49713592 | 0.14 |
ENSDART00000189624
|
BX323861.3
|
|
| chr2_-_10877765 | 0.14 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr13_+_2523032 | 0.14 |
ENSDART00000172261
|
lhb
|
luteinizing hormone, beta polypeptide |
| chr8_-_10048059 | 0.13 |
ENSDART00000138411
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr10_-_44467384 | 0.13 |
ENSDART00000186382
|
CABZ01072089.1
|
|
| chr8_-_13029297 | 0.13 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr22_+_20710434 | 0.13 |
ENSDART00000135521
|
peak3
|
PEAK family member 3 |
| chr18_-_36066087 | 0.13 |
ENSDART00000059352
ENSDART00000145177 |
exosc5
|
exosome component 5 |
| chr22_-_17671348 | 0.13 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr22_-_10165446 | 0.12 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr4_+_8680767 | 0.12 |
ENSDART00000182726
|
adipor2
|
adiponectin receptor 2 |
| chr9_+_54679221 | 0.12 |
ENSDART00000167769
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr25_-_16768078 | 0.12 |
ENSDART00000025186
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr12_-_44016898 | 0.12 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr3_+_60761811 | 0.12 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr7_-_17570923 | 0.12 |
ENSDART00000188476
ENSDART00000080624 |
nitr5
|
novel immune-type receptor 5 |
| chr10_+_14488625 | 0.12 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr6_-_52788213 | 0.12 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr7_+_12976173 | 0.12 |
ENSDART00000193150
ENSDART00000149518 |
slc25a22b
|
solute carrier family 25 member 22b |
| chr20_-_40760217 | 0.12 |
ENSDART00000101007
|
cx34.5
|
connexin 34.5 |
| chr15_+_846768 | 0.12 |
ENSDART00000155633
ENSDART00000191235 |
si:dkey-7i4.5
si:dkey-77f5.8
|
si:dkey-7i4.5 si:dkey-77f5.8 |
| chr1_+_40801448 | 0.12 |
ENSDART00000187394
|
cpz
|
carboxypeptidase Z |
| chr25_+_4787607 | 0.12 |
ENSDART00000159422
|
myo5c
|
myosin VC |
| chr22_-_9736050 | 0.11 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr15_+_25158104 | 0.11 |
ENSDART00000128267
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr9_+_29548195 | 0.11 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr2_+_44977889 | 0.11 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr8_+_22359881 | 0.11 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr15_+_47440477 | 0.11 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr3_-_55121125 | 0.11 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr7_-_28658143 | 0.11 |
ENSDART00000173556
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr1_-_17715493 | 0.11 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr5_-_24238733 | 0.11 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr20_+_46206998 | 0.11 |
ENSDART00000074482
|
taar15
|
trace amine associated receptor 15 |
| chr5_+_57480014 | 0.11 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr25_+_8407892 | 0.11 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr3_+_60277300 | 0.11 |
ENSDART00000170977
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr16_+_5908483 | 0.11 |
ENSDART00000167393
|
ulk4
|
unc-51 like kinase 4 |
| chr18_-_27897217 | 0.11 |
ENSDART00000175259
|
iqcg
|
IQ motif containing G |
| chr23_-_1658980 | 0.11 |
ENSDART00000186227
|
CU693481.1
|
|
| chr2_+_11119315 | 0.11 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr15_-_563877 | 0.11 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr14_+_33049579 | 0.11 |
ENSDART00000113470
|
tex11
|
testis expressed 11 |
| chr19_+_19747430 | 0.10 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr4_-_16658514 | 0.10 |
ENSDART00000133837
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr21_-_11646878 | 0.10 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr24_-_40700596 | 0.10 |
ENSDART00000162635
|
smyhc2
|
slow myosin heavy chain 2 |
| chr7_+_8358547 | 0.10 |
ENSDART00000173016
|
jac7
|
jacalin 7 |
| chr3_-_32817274 | 0.10 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr4_-_32192868 | 0.10 |
ENSDART00000164675
|
CR450785.2
|
|
| chr17_-_50040927 | 0.10 |
ENSDART00000184304
|
FO834825.1
|
|
| chr16_+_11660839 | 0.10 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr4_-_14954029 | 0.10 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr6_-_2134581 | 0.10 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr19_-_3773905 | 0.10 |
ENSDART00000168433
|
btr20
|
bloodthirsty-related gene family, member 20 |
| chr10_+_38708099 | 0.10 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr10_+_36294641 | 0.10 |
ENSDART00000158983
|
or106-8
|
odorant receptor, family G, subfamily 106, member 8 |
| chr14_+_12316581 | 0.10 |
ENSDART00000170115
ENSDART00000149757 |
si:ch211-125c23.3
cx31.7
|
si:ch211-125c23.3 connexin 31.7 |
| chr5_-_4216324 | 0.10 |
ENSDART00000168610
|
si:ch211-283g2.2
|
si:ch211-283g2.2 |
| chr5_+_38685089 | 0.10 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr4_+_42175261 | 0.10 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr23_+_7379728 | 0.10 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr3_-_46403778 | 0.10 |
ENSDART00000074422
|
cdip1
|
cell death-inducing p53 target 1 |
| chr7_+_66822229 | 0.10 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr9_+_1139378 | 0.10 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr5_-_4239765 | 0.09 |
ENSDART00000180015
|
si:ch211-283g2.3
|
si:ch211-283g2.3 |
| chr4_-_14954327 | 0.09 |
ENSDART00000182729
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr25_+_30074947 | 0.09 |
ENSDART00000154849
|
si:ch211-147k10.5
|
si:ch211-147k10.5 |
| chr4_+_71401321 | 0.09 |
ENSDART00000158076
|
si:ch211-76m11.7
|
si:ch211-76m11.7 |
| chr25_+_23280220 | 0.09 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr19_+_7810028 | 0.09 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr10_-_34223567 | 0.09 |
ENSDART00000184889
|
pimr145
|
Pim proto-oncogene, serine/threonine kinase, related 145 |
| chr12_+_2428247 | 0.09 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr25_+_25438322 | 0.09 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr1_+_55239710 | 0.09 |
ENSDART00000174846
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr10_-_24784446 | 0.09 |
ENSDART00000140877
|
si:ch1073-15f19.2
|
si:ch1073-15f19.2 |
| chr13_-_33321541 | 0.09 |
ENSDART00000144196
|
si:dkey-71p21.13
|
si:dkey-71p21.13 |
| chr11_+_36243774 | 0.09 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr1_+_9082417 | 0.09 |
ENSDART00000146303
|
cacng3a
|
calcium channel, voltage-dependent, gamma subunit 3a |
| chr19_-_2876321 | 0.09 |
ENSDART00000159253
|
exosc7
|
exosome component 7 |
| chr7_-_2309236 | 0.09 |
ENSDART00000173356
|
si:dkey-187j14.8
|
si:dkey-187j14.8 |
| chr13_+_48359573 | 0.09 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr22_-_11661724 | 0.09 |
ENSDART00000025198
|
mettl21a
|
methyltransferase like 21A |
| chr8_-_13419049 | 0.09 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr10_-_14488472 | 0.09 |
ENSDART00000101298
ENSDART00000138161 |
galt
|
galactose-1-phosphate uridylyltransferase |
| chr16_-_28658341 | 0.09 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr22_+_12595144 | 0.09 |
ENSDART00000140054
ENSDART00000060979 ENSDART00000139826 |
zgc:92335
|
zgc:92335 |
| chr8_-_13471916 | 0.09 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr21_-_20341836 | 0.09 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.4 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.1 | 0.3 | GO:0051000 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.1 | 0.3 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.2 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0097201 | response to acidic pH(GO:0010447) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.4 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.4 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.2 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.2 | GO:0035545 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.0 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 1.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0021703 | locus ceruleus development(GO:0021703) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.0 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |