Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
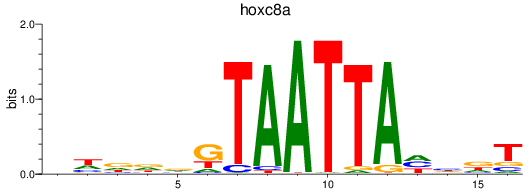
Results for hoxc8a
Z-value: 0.34
Transcription factors associated with hoxc8a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc8a
|
ENSDARG00000070346 | homeobox C8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc8a | dr11_v1_chr23_+_36101185_36101185 | -0.09 | 8.9e-01 | Click! |
Activity profile of hoxc8a motif
Sorted Z-values of hoxc8a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_23298928 | 0.33 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr4_-_815871 | 0.31 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr15_-_15357178 | 0.31 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr11_-_1509773 | 0.29 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr13_-_35808904 | 0.28 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr18_+_7283283 | 0.27 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr20_+_40457599 | 0.27 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr13_+_13693722 | 0.27 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr5_+_27137473 | 0.26 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr15_+_15856178 | 0.26 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr2_+_50608099 | 0.26 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr23_-_29505645 | 0.25 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr8_+_31717175 | 0.25 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr13_+_38521152 | 0.24 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr5_+_30635309 | 0.24 |
ENSDART00000183769
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr8_+_31716872 | 0.23 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr2_-_16380283 | 0.22 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr16_+_17389116 | 0.20 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr2_+_31833997 | 0.19 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr14_-_30587814 | 0.19 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr4_+_76926059 | 0.19 |
ENSDART00000136192
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr5_+_36768674 | 0.19 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr6_-_30210378 | 0.18 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr8_+_18624658 | 0.18 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr18_+_924949 | 0.18 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr20_-_14925281 | 0.18 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr6_+_40591149 | 0.17 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr3_+_46628885 | 0.17 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr16_-_44945224 | 0.17 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr14_-_47248784 | 0.17 |
ENSDART00000135479
|
fstl5
|
follistatin-like 5 |
| chr14_-_27121854 | 0.16 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr2_-_33687214 | 0.16 |
ENSDART00000147439
|
atp6v0b
|
ATPase H+ transporting V0 subunit b |
| chr18_+_27738349 | 0.16 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr16_+_39159752 | 0.15 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr17_+_23554932 | 0.15 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr20_-_14924858 | 0.15 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr13_+_12299997 | 0.14 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr2_-_30668580 | 0.14 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr19_+_5480327 | 0.13 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr17_+_26611929 | 0.13 |
ENSDART00000166450
ENSDART00000087023 |
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr23_+_40604951 | 0.13 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr24_-_29963858 | 0.13 |
ENSDART00000183442
|
CR352310.1
|
|
| chr11_-_6188413 | 0.12 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr17_-_43556208 | 0.12 |
ENSDART00000188424
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr7_-_67894046 | 0.12 |
ENSDART00000168964
|
si:ch73-315f9.2
|
si:ch73-315f9.2 |
| chr18_+_17827149 | 0.12 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr20_+_38201644 | 0.11 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr17_-_43556415 | 0.11 |
ENSDART00000190102
ENSDART00000193156 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr20_-_9462433 | 0.11 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr24_-_7321928 | 0.11 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr15_-_35252522 | 0.11 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr21_-_34032650 | 0.11 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr16_+_13818743 | 0.10 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr21_-_5856050 | 0.10 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr25_-_13490744 | 0.10 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr15_-_42736433 | 0.10 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr11_+_11267493 | 0.10 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr20_-_34750045 | 0.10 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr5_-_68093169 | 0.10 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr13_-_39736938 | 0.09 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr4_+_14727018 | 0.09 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chrM_+_9052 | 0.09 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr11_-_40728380 | 0.09 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr20_-_34750363 | 0.08 |
ENSDART00000152845
|
znf395b
|
zinc finger protein 395b |
| chr13_-_31812394 | 0.08 |
ENSDART00000124445
|
sertad4
|
SERTA domain containing 4 |
| chr15_-_14552101 | 0.08 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr23_-_35790235 | 0.07 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr7_-_46777876 | 0.07 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr15_+_29140126 | 0.07 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr11_+_33818179 | 0.07 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr25_-_7705487 | 0.07 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr9_-_21918963 | 0.07 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr19_-_205104 | 0.07 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr11_-_21528056 | 0.06 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr8_-_7567815 | 0.06 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr4_+_14727212 | 0.06 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr7_-_12464412 | 0.06 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr6_-_40713183 | 0.06 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr3_+_22442445 | 0.06 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr1_+_22691256 | 0.06 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr1_-_58036509 | 0.06 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr21_+_22738939 | 0.06 |
ENSDART00000151342
ENSDART00000079145 |
arhgap42a
|
Rho GTPase activating protein 42a |
| chr13_+_36622100 | 0.06 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr5_+_63857055 | 0.05 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr15_-_23420079 | 0.05 |
ENSDART00000104525
ENSDART00000148840 |
kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr21_+_29077509 | 0.05 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr2_+_16482338 | 0.05 |
ENSDART00000143912
|
fbxo36b
|
F-box protein 36b |
| chr13_+_31070181 | 0.05 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr7_-_48667056 | 0.05 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr1_+_39482588 | 0.05 |
ENSDART00000181790
|
tenm3
|
teneurin transmembrane protein 3 |
| chr15_+_31899312 | 0.05 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr21_+_30355767 | 0.05 |
ENSDART00000189948
|
CR749164.1
|
|
| chr7_-_32895668 | 0.05 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr10_-_5847655 | 0.04 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr6_+_49722970 | 0.04 |
ENSDART00000155934
ENSDART00000154738 |
stx16
|
syntaxin 16 |
| chrM_+_9735 | 0.04 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr16_-_13623928 | 0.04 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr6_+_19383267 | 0.04 |
ENSDART00000166549
|
mchr1a
|
melanin-concentrating hormone receptor 1a |
| chr4_+_49506359 | 0.04 |
ENSDART00000154478
|
si:dkey-5i16.5
|
si:dkey-5i16.5 |
| chr11_+_30244356 | 0.04 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr10_+_2899108 | 0.04 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr23_+_36130883 | 0.04 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr21_+_8427059 | 0.04 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr7_-_30174882 | 0.04 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr11_-_44979281 | 0.04 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr22_+_29113796 | 0.04 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr4_+_5333988 | 0.04 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr6_+_49723289 | 0.03 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr5_+_69981453 | 0.03 |
ENSDART00000143860
|
si:ch211-154e10.1
|
si:ch211-154e10.1 |
| chr16_-_13623659 | 0.03 |
ENSDART00000168978
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr24_+_26997798 | 0.03 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr8_+_16676894 | 0.03 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr2_+_20793982 | 0.03 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr19_+_33850705 | 0.03 |
ENSDART00000160356
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr10_-_26744131 | 0.03 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr5_+_29671681 | 0.03 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr13_+_35339182 | 0.03 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr20_-_27225876 | 0.03 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr10_+_11355841 | 0.02 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr1_+_524388 | 0.02 |
ENSDART00000020327
|
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr17_+_50248152 | 0.02 |
ENSDART00000153998
|
zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr9_+_32178374 | 0.02 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr19_-_33850201 | 0.02 |
ENSDART00000168583
|
otud6b
|
OTU domain containing 6B |
| chr21_+_35327589 | 0.02 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr15_-_38154616 | 0.02 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr13_-_45811137 | 0.02 |
ENSDART00000190512
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr10_-_35108683 | 0.01 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr2_-_28671139 | 0.01 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr21_+_35328025 | 0.01 |
ENSDART00000136211
|
rars
|
arginyl-tRNA synthetase |
| chr24_+_36204028 | 0.01 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr3_+_7808459 | 0.01 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr24_+_22485710 | 0.01 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr11_+_45110865 | 0.01 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr3_+_30500968 | 0.00 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr3_+_29179329 | 0.00 |
ENSDART00000085216
ENSDART00000190136 |
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr14_-_16082806 | 0.00 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr8_-_39822917 | 0.00 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr3_-_34084387 | 0.00 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr16_+_4497302 | 0.00 |
ENSDART00000081826
ENSDART00000148096 |
ttc29
|
tetratricopeptide repeat domain 29 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc8a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0033344 | cholesterol efflux(GO:0033344) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |