Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
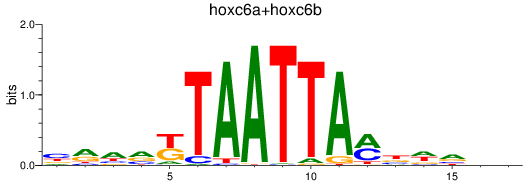
Results for hoxc6a+hoxc6b
Z-value: 0.51
Transcription factors associated with hoxc6a+hoxc6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc6a
|
ENSDARG00000070343 | homeobox C6a |
|
hoxc6b
|
ENSDARG00000101954 | homeobox C6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc6b | dr11_v1_chr11_+_2198831_2198831 | -0.37 | 5.4e-01 | Click! |
| hoxc6a | dr11_v1_chr23_+_36115541_36115541 | -0.23 | 7.0e-01 | Click! |
Activity profile of hoxc6a+hoxc6b motif
Sorted Z-values of hoxc6a+hoxc6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24791505 | 0.57 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr18_+_13162728 | 0.55 |
ENSDART00000101472
|
tat
|
tyrosine aminotransferase |
| chr2_+_56657804 | 0.42 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr20_+_54299419 | 0.41 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr24_-_38110779 | 0.40 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr20_+_54309148 | 0.40 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr20_+_54304800 | 0.37 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr22_-_16154771 | 0.30 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr14_-_46173265 | 0.30 |
ENSDART00000164321
|
zmp:0000000758
|
zmp:0000000758 |
| chr2_-_17392799 | 0.27 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr15_-_46779934 | 0.26 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr5_-_50992690 | 0.23 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr25_+_22320738 | 0.22 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr21_+_45841731 | 0.20 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr2_-_17393216 | 0.19 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr7_+_24573721 | 0.18 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr1_-_513762 | 0.18 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr2_-_31833347 | 0.18 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr6_-_12588044 | 0.18 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr6_+_25257728 | 0.17 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr24_+_16985181 | 0.16 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr3_-_59297532 | 0.15 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr10_-_34916208 | 0.15 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr11_-_10456553 | 0.15 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr22_+_19552987 | 0.15 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr4_+_57881965 | 0.15 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr11_-_10456387 | 0.14 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr13_-_52089003 | 0.14 |
ENSDART00000187600
|
tmem254
|
transmembrane protein 254 |
| chr5_+_71802014 | 0.14 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr13_+_646700 | 0.12 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr22_-_36530902 | 0.12 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr6_-_52788213 | 0.12 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr20_+_38032143 | 0.12 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr20_-_5291012 | 0.12 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr2_+_20406399 | 0.12 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr2_-_10877765 | 0.12 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr1_-_9195629 | 0.11 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr17_+_16564921 | 0.11 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr8_-_53044300 | 0.11 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr17_+_51682429 | 0.10 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr23_-_19140781 | 0.10 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr7_-_8712148 | 0.10 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr12_+_16168342 | 0.10 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr23_+_1029450 | 0.10 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr24_-_38657683 | 0.10 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr19_-_47323267 | 0.09 |
ENSDART00000190077
|
CU138512.1
|
|
| chr5_-_42904329 | 0.09 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr7_+_4694924 | 0.09 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr15_-_17618800 | 0.09 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr1_+_49668423 | 0.09 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr7_-_66868543 | 0.09 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr7_+_4694762 | 0.09 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr2_-_23603061 | 0.09 |
ENSDART00000132072
|
si:dkey-58b18.9
|
si:dkey-58b18.9 |
| chr1_+_51721851 | 0.09 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr3_+_46635527 | 0.09 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr3_+_23092762 | 0.09 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr5_-_37103487 | 0.08 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr2_+_14992879 | 0.08 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr21_-_1640547 | 0.08 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr10_+_29850330 | 0.08 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr9_+_54290896 | 0.08 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr25_-_37186894 | 0.08 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr5_+_19343880 | 0.08 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr10_-_13343831 | 0.08 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr9_+_11281969 | 0.08 |
ENSDART00000110691
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr22_-_9183944 | 0.08 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr4_-_19921410 | 0.08 |
ENSDART00000184359
ENSDART00000179965 |
BX323820.1
|
|
| chr15_-_17619306 | 0.08 |
ENSDART00000184011
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr16_+_29509133 | 0.08 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr4_-_11064073 | 0.08 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr10_+_31248036 | 0.08 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr5_+_55225497 | 0.08 |
ENSDART00000144087
|
tmc2a
|
transmembrane channel-like 2a |
| chr22_-_27296889 | 0.08 |
ENSDART00000155724
|
si:dkey-208m12.3
|
si:dkey-208m12.3 |
| chr3_+_23029484 | 0.08 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr25_+_29474982 | 0.08 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr2_-_15324837 | 0.08 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr13_+_9468535 | 0.08 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr9_+_41080029 | 0.08 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr7_+_4702269 | 0.08 |
ENSDART00000139880
|
si:ch211-225k7.4
|
si:ch211-225k7.4 |
| chr10_-_34046757 | 0.08 |
ENSDART00000099648
|
pimr149
|
Pim proto-oncogene, serine/threonine kinase, related 149 |
| chr12_-_35944654 | 0.08 |
ENSDART00000162579
ENSDART00000164199 |
dnai2a
|
dynein, axonemal, intermediate chain 2a |
| chr21_-_44081540 | 0.07 |
ENSDART00000130833
|
FO704810.1
|
|
| chr13_-_9467944 | 0.07 |
ENSDART00000136582
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr3_-_50443607 | 0.07 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr8_+_29635968 | 0.07 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr2_+_3201345 | 0.07 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr21_-_26028205 | 0.07 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr17_+_30369396 | 0.07 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr22_+_1568306 | 0.07 |
ENSDART00000163223
|
si:ch211-255f4.9
|
si:ch211-255f4.9 |
| chr22_-_5252005 | 0.07 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr6_-_16717878 | 0.07 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr7_+_4657987 | 0.07 |
ENSDART00000144321
ENSDART00000142344 |
si:dkey-83f18.2
|
si:dkey-83f18.2 |
| chr13_-_50366950 | 0.06 |
ENSDART00000098209
|
sirt1
|
sirtuin 1 |
| chr2_+_19522082 | 0.06 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr10_+_34047352 | 0.06 |
ENSDART00000169333
|
HTRA2 (1 of many)
|
si:dkey-10b15.8 |
| chr4_+_17279966 | 0.06 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr6_+_7533601 | 0.06 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr12_-_4532066 | 0.06 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr5_+_66433287 | 0.06 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr12_+_47698356 | 0.06 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr20_+_25586099 | 0.06 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr4_-_43640507 | 0.06 |
ENSDART00000150700
|
si:dkey-29p23.2
|
si:dkey-29p23.2 |
| chr19_-_5769553 | 0.06 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr24_+_13316737 | 0.06 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr14_+_36521005 | 0.06 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr15_+_34592215 | 0.06 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr21_-_26490186 | 0.06 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr5_+_65871688 | 0.05 |
ENSDART00000175217
|
FAM163B (1 of many)
|
family with sequence similarity 163 member B |
| chr19_-_5769728 | 0.05 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr3_+_7808459 | 0.05 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr11_-_11266882 | 0.05 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr3_-_13921173 | 0.05 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr17_+_20589553 | 0.05 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr13_+_35528607 | 0.05 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr23_-_18024543 | 0.05 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr2_+_20605925 | 0.05 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr3_+_40164129 | 0.05 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr11_+_34760628 | 0.05 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr1_+_58242498 | 0.04 |
ENSDART00000149091
|
ggt1l2.2
|
gamma-glutamyltransferase 1 like 2.2 |
| chr24_+_21514283 | 0.04 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr2_-_17235891 | 0.04 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr2_+_10878406 | 0.04 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr14_-_48765262 | 0.04 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr5_+_12888260 | 0.04 |
ENSDART00000175916
|
gal3st1a
|
galactose-3-O-sulfotransferase 1a |
| chr3_+_29179329 | 0.04 |
ENSDART00000085216
ENSDART00000190136 |
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr11_+_11974708 | 0.04 |
ENSDART00000125060
|
zgc:64002
|
zgc:64002 |
| chr14_+_46313396 | 0.04 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr3_+_38540411 | 0.04 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr7_+_7696665 | 0.04 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr24_+_34113424 | 0.04 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr8_+_36560019 | 0.04 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr22_-_10121880 | 0.04 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr8_-_39884359 | 0.04 |
ENSDART00000131372
|
mlec
|
malectin |
| chr21_+_43172506 | 0.04 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr9_+_310331 | 0.04 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr2_+_19578079 | 0.04 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr11_+_1602916 | 0.04 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr3_-_31158382 | 0.04 |
ENSDART00000076764
ENSDART00000076796 |
smg1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr25_-_21031007 | 0.04 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr20_-_45812144 | 0.04 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr4_+_59845617 | 0.03 |
ENSDART00000167626
ENSDART00000123157 |
si:dkey-196n19.2
|
si:dkey-196n19.2 |
| chr17_+_12285285 | 0.03 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr18_+_20481982 | 0.03 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr18_+_34225520 | 0.03 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr19_+_10592778 | 0.03 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr24_+_19415124 | 0.03 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr15_+_857148 | 0.03 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr21_+_25236297 | 0.03 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr16_-_27677930 | 0.03 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr12_-_28363111 | 0.03 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr8_+_52637507 | 0.03 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr22_+_508290 | 0.03 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr13_-_39736938 | 0.03 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr1_+_32051581 | 0.03 |
ENSDART00000146602
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr3_-_31254379 | 0.03 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr6_-_43283122 | 0.03 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr19_+_1688727 | 0.03 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr18_+_13275735 | 0.02 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr1_-_22757145 | 0.02 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr1_-_42289704 | 0.02 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr4_-_49582758 | 0.02 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr3_+_1637358 | 0.02 |
ENSDART00000180266
|
CR394546.5
|
|
| chr20_-_38617766 | 0.02 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr7_+_59020972 | 0.02 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr5_-_13251907 | 0.02 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr11_-_29737088 | 0.02 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr20_-_9095105 | 0.02 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr6_+_24398907 | 0.02 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr5_-_28109416 | 0.02 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr10_-_5847655 | 0.01 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr4_-_43388943 | 0.01 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr21_-_36571804 | 0.01 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr5_-_51198430 | 0.01 |
ENSDART00000132503
ENSDART00000097473 ENSDART00000165870 |
snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr16_-_41515662 | 0.01 |
ENSDART00000166201
ENSDART00000127243 |
siglec15l
|
sialic acid binding Ig-like lectin 15, like |
| chr24_+_9693951 | 0.01 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr19_+_7847920 | 0.01 |
ENSDART00000132454
|
si:dkeyp-85e10.1
|
si:dkeyp-85e10.1 |
| chr4_-_75158035 | 0.01 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr20_-_13660767 | 0.01 |
ENSDART00000127654
|
TAGAP
|
si:ch211-122h15.4 |
| chr7_-_11596450 | 0.01 |
ENSDART00000173863
|
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr20_+_25225112 | 0.01 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr4_-_5019113 | 0.01 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr20_-_35040041 | 0.00 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr16_-_7793457 | 0.00 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr4_-_5108844 | 0.00 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr14_+_4807207 | 0.00 |
ENSDART00000167145
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr15_+_31899312 | 0.00 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc6a+hoxc6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.1 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |