Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
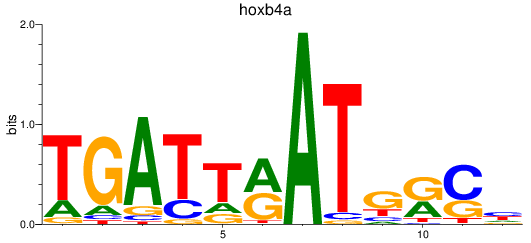
Results for hoxb4a
Z-value: 0.87
Transcription factors associated with hoxb4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb4a
|
ENSDARG00000013533 | homeobox B4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb4a | dr11_v1_chr3_+_23710839_23710839 | -0.94 | 1.8e-02 | Click! |
Activity profile of hoxb4a motif
Sorted Z-values of hoxb4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_20901505 | 0.92 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr17_+_23937262 | 0.56 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr20_-_29420713 | 0.49 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr25_-_4482449 | 0.46 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr21_-_24632778 | 0.43 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr24_+_32472155 | 0.41 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr25_-_13842618 | 0.40 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr10_-_20445549 | 0.40 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr18_-_14836600 | 0.40 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr12_+_9703172 | 0.39 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr7_-_41964877 | 0.38 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr18_-_12957451 | 0.38 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr18_-_42172101 | 0.36 |
ENSDART00000124211
|
cntn5
|
contactin 5 |
| chr18_-_14836862 | 0.35 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr17_+_27434626 | 0.35 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr2_-_54039293 | 0.34 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr2_+_27010439 | 0.34 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr6_+_18367388 | 0.33 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr16_-_32672883 | 0.32 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr7_-_19369002 | 0.32 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr11_-_13341483 | 0.25 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr19_+_14059349 | 0.25 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr23_-_33038423 | 0.25 |
ENSDART00000180539
|
plxna2
|
plexin A2 |
| chr11_-_13341051 | 0.24 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr2_+_37480669 | 0.24 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr7_-_52842007 | 0.24 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr17_+_19626479 | 0.23 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr1_-_681116 | 0.23 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr9_-_3653259 | 0.23 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr5_-_18446483 | 0.22 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr10_-_26744131 | 0.22 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr17_-_8727699 | 0.21 |
ENSDART00000049236
ENSDART00000149505 ENSDART00000148619 ENSDART00000149668 ENSDART00000148827 |
ctbp2a
|
C-terminal binding protein 2a |
| chr12_+_28854410 | 0.21 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr19_-_10425140 | 0.21 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr3_-_52644737 | 0.18 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr9_-_15789526 | 0.18 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr15_-_26552652 | 0.17 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr1_-_46981134 | 0.17 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr13_-_33207367 | 0.16 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr6_-_29105727 | 0.16 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr21_-_20328375 | 0.16 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr11_+_13223625 | 0.16 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr18_+_6126506 | 0.16 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr5_-_7834582 | 0.15 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr23_-_3721444 | 0.13 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr20_+_51479263 | 0.13 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr2_-_36818132 | 0.12 |
ENSDART00000110447
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr23_+_13814978 | 0.12 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr6_-_57635577 | 0.11 |
ENSDART00000168708
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr20_+_51478939 | 0.11 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr20_-_32188897 | 0.11 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr1_+_1712140 | 0.11 |
ENSDART00000081047
|
atp1a1a.1
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 1 |
| chr12_+_28854963 | 0.11 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr24_-_26460853 | 0.10 |
ENSDART00000078628
|
slc7a14b
|
solute carrier family 7, member 14b |
| chr4_-_17257435 | 0.10 |
ENSDART00000131973
|
lrmp
|
lymphoid-restricted membrane protein |
| chr2_+_44972720 | 0.10 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr14_-_30876708 | 0.10 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr3_-_51109286 | 0.09 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr9_+_16449398 | 0.09 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr23_+_27756984 | 0.08 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr16_-_18960613 | 0.08 |
ENSDART00000183197
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr21_+_22423286 | 0.07 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr15_+_25489406 | 0.07 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr14_+_46274611 | 0.06 |
ENSDART00000134363
|
cabp2b
|
calcium binding protein 2b |
| chr3_+_33300522 | 0.06 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr24_+_26402110 | 0.06 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr14_-_30876299 | 0.05 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr11_+_40530748 | 0.05 |
ENSDART00000173239
|
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr4_-_13255700 | 0.05 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr13_+_36622100 | 0.05 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr1_+_11881559 | 0.05 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr6_-_28345002 | 0.03 |
ENSDART00000158955
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr13_+_30912117 | 0.03 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr2_-_57473980 | 0.02 |
ENSDART00000149353
ENSDART00000150034 |
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr5_-_64103863 | 0.01 |
ENSDART00000135014
ENSDART00000083684 |
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr13_+_30912385 | 0.01 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr1_-_11372456 | 0.01 |
ENSDART00000144164
ENSDART00000141238 |
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr18_-_25401002 | 0.00 |
ENSDART00000055567
|
gnrhr4
|
gonadotropin releasing hormone receptor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.5 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.1 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |