Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
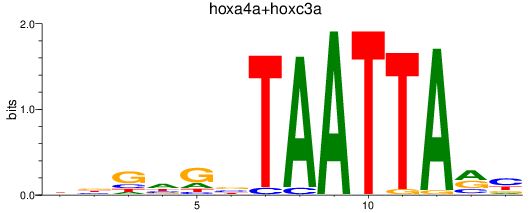
Results for hoxa4a+hoxc3a
Z-value: 0.59
Transcription factors associated with hoxa4a+hoxc3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc3a
|
ENSDARG00000070339 | homeobox C3a |
|
hoxa4a
|
ENSDARG00000103862 | homeobox A4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa3a | dr11_v1_chr19_+_19767567_19767567 | -0.65 | 2.4e-01 | Click! |
| hoxc3a | dr11_v1_chr23_+_36106790_36106790 | 0.22 | 7.3e-01 | Click! |
Activity profile of hoxa4a+hoxc3a motif
Sorted Z-values of hoxa4a+hoxc3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_5369486 | 0.49 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr20_-_37813863 | 0.47 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_+_35075847 | 0.41 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr6_-_54815886 | 0.37 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr12_+_22580579 | 0.37 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_-_5358443 | 0.35 |
ENSDART00000105036
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr4_+_11464255 | 0.34 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr11_-_20096018 | 0.33 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr15_-_47848544 | 0.33 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr21_-_42876565 | 0.31 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr23_+_20110086 | 0.27 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr10_-_22095505 | 0.26 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr22_-_30770751 | 0.24 |
ENSDART00000172115
|
AL831726.2
|
|
| chr24_-_36680261 | 0.23 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr21_+_25777425 | 0.22 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr20_+_53577502 | 0.22 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr21_-_26490186 | 0.21 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr25_+_37293312 | 0.21 |
ENSDART00000086737
ENSDART00000161595 |
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr20_-_9462433 | 0.20 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr1_-_23308225 | 0.19 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr19_+_43297546 | 0.19 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr14_-_4145594 | 0.19 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr16_-_51271962 | 0.19 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr12_+_20352400 | 0.19 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr3_-_16784280 | 0.19 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr22_+_6471584 | 0.19 |
ENSDART00000133974
|
si:ch211-105n15.1
|
si:ch211-105n15.1 |
| chr24_-_33291784 | 0.19 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr4_+_77943184 | 0.18 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr7_+_4474880 | 0.18 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr8_-_31107537 | 0.18 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr3_-_15734358 | 0.17 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr20_+_36812368 | 0.17 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr19_+_823945 | 0.17 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr19_+_2631565 | 0.16 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr12_-_46176115 | 0.16 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr6_-_7720332 | 0.16 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr6_+_16468776 | 0.16 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr12_-_35830625 | 0.16 |
ENSDART00000180028
|
CU459056.1
|
|
| chr7_-_71389375 | 0.15 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr4_+_6032640 | 0.15 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr17_-_37395460 | 0.15 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr3_-_15734530 | 0.15 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr24_-_27452488 | 0.15 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr13_+_45431660 | 0.15 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr14_+_3038473 | 0.15 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr5_+_38623814 | 0.15 |
ENSDART00000146046
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr17_-_20118145 | 0.15 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr4_+_73085993 | 0.15 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr7_+_31891110 | 0.14 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr8_-_25034411 | 0.14 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr24_-_37640705 | 0.14 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr4_-_9891874 | 0.14 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr10_-_21362071 | 0.13 |
ENSDART00000125167
|
avd
|
avidin |
| chr14_-_1355544 | 0.13 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr24_+_22731228 | 0.13 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr6_-_55399214 | 0.13 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr12_-_46112892 | 0.13 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr17_+_16046314 | 0.13 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_-_42904329 | 0.13 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr10_-_2971407 | 0.13 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr25_+_36292057 | 0.12 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr10_+_39212898 | 0.12 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr24_-_6078222 | 0.12 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr19_-_44064842 | 0.12 |
ENSDART00000151541
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr3_+_27798094 | 0.12 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr16_-_20312146 | 0.12 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr11_+_31609481 | 0.11 |
ENSDART00000124830
ENSDART00000162768 |
zgc:162816
|
zgc:162816 |
| chr16_+_42471455 | 0.11 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr25_+_5035343 | 0.11 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr10_-_21362320 | 0.11 |
ENSDART00000189789
|
avd
|
avidin |
| chr6_-_43283122 | 0.11 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr22_+_6254194 | 0.11 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr18_+_27571448 | 0.11 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr23_-_36446307 | 0.11 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr18_-_46258612 | 0.11 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr14_-_7207961 | 0.11 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr4_+_35594200 | 0.11 |
ENSDART00000193902
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr12_-_4532066 | 0.11 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr18_+_23408073 | 0.11 |
ENSDART00000136489
|
mctp2a
|
multiple C2 domains, transmembrane 2a |
| chr22_+_19366866 | 0.11 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr3_-_59297532 | 0.10 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr20_-_49704915 | 0.10 |
ENSDART00000189232
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr17_+_16046132 | 0.10 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr2_+_36112273 | 0.10 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr16_+_23799622 | 0.10 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr16_+_28994709 | 0.10 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr16_-_11798994 | 0.10 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr10_+_28160265 | 0.10 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr14_-_33481428 | 0.10 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr13_+_33688474 | 0.10 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr13_-_2010191 | 0.10 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr21_+_15592426 | 0.10 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr12_-_48188928 | 0.10 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr21_-_45728069 | 0.10 |
ENSDART00000185101
|
CU302316.1
|
|
| chr5_-_25733745 | 0.10 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr3_-_31079186 | 0.09 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr23_-_7674902 | 0.09 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr9_-_27398369 | 0.09 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr6_-_48418021 | 0.09 |
ENSDART00000090528
|
rhoca
|
ras homolog family member Ca |
| chr2_-_59247811 | 0.09 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr5_+_37068223 | 0.09 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr13_+_27232848 | 0.09 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr11_-_1550709 | 0.09 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr9_+_492459 | 0.09 |
ENSDART00000112635
|
CU984600.1
|
|
| chr1_-_45616470 | 0.09 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_+_513986 | 0.09 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr2_-_26596794 | 0.09 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr22_+_18156000 | 0.09 |
ENSDART00000143483
ENSDART00000136133 |
nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr4_+_31405646 | 0.09 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr15_+_21262917 | 0.09 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr18_+_924949 | 0.09 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr20_+_29209767 | 0.09 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr15_-_46779934 | 0.09 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr19_-_26923273 | 0.09 |
ENSDART00000089609
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr11_-_15296805 | 0.09 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr21_+_23135322 | 0.09 |
ENSDART00000159586
ENSDART00000125907 |
htr3a
|
5-hydroxytryptamine (serotonin) receptor 3A |
| chr19_-_4793263 | 0.09 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr10_+_16584382 | 0.09 |
ENSDART00000112039
|
CR790388.1
|
|
| chr21_-_37075542 | 0.09 |
ENSDART00000129439
|
BX649250.1
|
|
| chr16_+_54875530 | 0.08 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr7_+_17816006 | 0.08 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr13_-_31017960 | 0.08 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr23_+_31107685 | 0.08 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr22_-_10121880 | 0.08 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr14_+_40874608 | 0.08 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr18_-_40708537 | 0.08 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr25_-_6292560 | 0.08 |
ENSDART00000153496
|
wdr61
|
WD repeat domain 61 |
| chr15_-_43284021 | 0.08 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr2_+_5371492 | 0.08 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr23_-_24394719 | 0.08 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr11_+_3254524 | 0.08 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr5_-_30074332 | 0.08 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr9_+_30421489 | 0.08 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr8_-_23780334 | 0.08 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr9_+_41080029 | 0.08 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr2_-_28671139 | 0.08 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr3_-_50443607 | 0.08 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr1_-_27014872 | 0.08 |
ENSDART00000147414
ENSDART00000134032 ENSDART00000192087 ENSDART00000189111 ENSDART00000187348 ENSDART00000187248 |
cntln
|
centlein, centrosomal protein |
| chr19_+_4800549 | 0.08 |
ENSDART00000056235
|
st3gal1l
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1, like |
| chr4_+_306036 | 0.08 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr24_-_25166720 | 0.07 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr18_+_26422124 | 0.07 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr1_+_44127292 | 0.07 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr4_-_13548806 | 0.07 |
ENSDART00000067155
|
il22
|
interleukin 22 |
| chr1_+_40199074 | 0.07 |
ENSDART00000179679
ENSDART00000136849 |
si:ch211-113e8.5
|
si:ch211-113e8.5 |
| chr6_-_31361546 | 0.07 |
ENSDART00000027550
|
ak4
|
adenylate kinase 4 |
| chr22_-_38621438 | 0.07 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr8_-_11988065 | 0.07 |
ENSDART00000005140
|
med27
|
mediator complex subunit 27 |
| chr25_+_469855 | 0.07 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr22_+_737211 | 0.07 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr8_+_20776654 | 0.07 |
ENSDART00000135850
|
nfic
|
nuclear factor I/C |
| chr1_-_51710225 | 0.07 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr19_+_26923274 | 0.07 |
ENSDART00000148439
ENSDART00000148877 |
nelfe
|
negative elongation factor complex member E |
| chr25_-_35136267 | 0.07 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr16_+_24733741 | 0.07 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr20_+_29209926 | 0.07 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr21_-_5799122 | 0.07 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr22_+_19407531 | 0.07 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr1_-_13989643 | 0.07 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr22_-_9677264 | 0.07 |
ENSDART00000181737
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr20_+_29209615 | 0.07 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr1_+_53842344 | 0.07 |
ENSDART00000114506
|
crfb17
|
cytokine receptor family member B17 |
| chr9_+_43799829 | 0.07 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr4_+_966061 | 0.07 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr5_+_66433287 | 0.06 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr22_-_6229275 | 0.06 |
ENSDART00000146045
ENSDART00000179730 |
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr21_-_13661631 | 0.06 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr15_+_12429206 | 0.06 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr6_+_41191482 | 0.06 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr22_-_7702958 | 0.06 |
ENSDART00000193046
|
si:ch211-59h6.1
|
si:ch211-59h6.1 |
| chr25_-_19608382 | 0.06 |
ENSDART00000022279
ENSDART00000135201 ENSDART00000147223 ENSDART00000190220 ENSDART00000184242 ENSDART00000166824 |
gtse1
|
G-2 and S-phase expressed 1 |
| chr11_+_12811906 | 0.06 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr10_+_7593185 | 0.06 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr6_+_7414215 | 0.06 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr9_+_37152564 | 0.06 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr24_-_7995748 | 0.06 |
ENSDART00000158566
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr1_+_10318089 | 0.06 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr24_+_37640626 | 0.06 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr14_-_9713549 | 0.06 |
ENSDART00000193356
ENSDART00000166739 |
si:zfos-2326c3.2
|
si:zfos-2326c3.2 |
| chr9_-_43644261 | 0.06 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr18_+_16940440 | 0.06 |
ENSDART00000172560
|
si:dkey-8l13.5
|
si:dkey-8l13.5 |
| chr6_-_59271270 | 0.06 |
ENSDART00000126870
|
ndufa4l2b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2b |
| chr2_+_8779164 | 0.06 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr9_-_3934963 | 0.06 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr1_-_55118745 | 0.06 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr14_+_16287968 | 0.06 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr8_-_52562672 | 0.06 |
ENSDART00000159333
ENSDART00000159974 |
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr17_+_8799451 | 0.06 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr14_-_413273 | 0.06 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr4_+_74929427 | 0.06 |
ENSDART00000174082
|
nup50
|
nucleoporin 50 |
| chr16_-_30932533 | 0.06 |
ENSDART00000085761
|
slc45a4
|
solute carrier family 45, member 4 |
| chr19_-_18152942 | 0.06 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr24_+_17260329 | 0.06 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr14_+_30795559 | 0.06 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr10_+_16036246 | 0.06 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr11_-_18020258 | 0.06 |
ENSDART00000156116
|
qrich1
|
glutamine-rich 1 |
| chr13_+_35339182 | 0.05 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr15_+_5360407 | 0.05 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr10_+_44700103 | 0.05 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr12_-_314899 | 0.05 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa4a+hoxc3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0045830 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.0 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.0 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.0 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.2 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |