Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
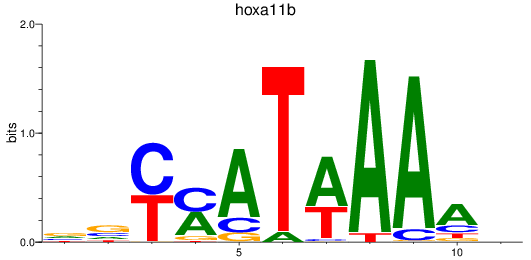
Results for hoxa11b
Z-value: 1.06
Transcription factors associated with hoxa11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11b
|
ENSDARG00000007009 | homeobox A11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11b | dr11_v1_chr16_+_20904754_20904754 | -0.13 | 8.3e-01 | Click! |
Activity profile of hoxa11b motif
Sorted Z-values of hoxa11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_36034582 | 0.43 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr19_-_47571456 | 0.37 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_+_5531138 | 0.37 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr13_-_37619159 | 0.32 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr24_-_10014512 | 0.30 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr21_+_11401247 | 0.28 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr16_-_45917322 | 0.28 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr3_+_46763745 | 0.28 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr10_-_8060573 | 0.28 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr9_+_52398531 | 0.26 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr2_-_44777592 | 0.25 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr24_-_38083378 | 0.24 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr5_-_42272517 | 0.24 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr22_+_6293563 | 0.24 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr10_+_26612321 | 0.23 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr3_-_32337653 | 0.23 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr20_+_26538137 | 0.23 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr20_-_29499363 | 0.23 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_+_46764022 | 0.23 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr12_-_25887864 | 0.22 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr8_+_52637507 | 0.22 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr12_-_4249000 | 0.22 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr5_-_4532516 | 0.22 |
ENSDART00000192398
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr3_+_62161184 | 0.21 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr11_-_19775182 | 0.21 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr22_-_24297510 | 0.21 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr17_+_53294228 | 0.21 |
ENSDART00000158172
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr5_+_9037650 | 0.21 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr12_-_5728755 | 0.20 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr21_-_7035599 | 0.20 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr22_-_24818066 | 0.19 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr7_-_23768234 | 0.19 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr22_-_10158038 | 0.19 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr3_-_19495814 | 0.19 |
ENSDART00000162248
|
CU571315.3
|
|
| chr3_+_32129632 | 0.19 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr21_+_20396858 | 0.19 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr3_-_26183699 | 0.19 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr13_+_27329556 | 0.19 |
ENSDART00000140085
|
mb21d1
|
Mab-21 domain containing 1 |
| chr9_-_2892250 | 0.19 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr23_-_43718067 | 0.19 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr9_-_48214216 | 0.18 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr14_-_4120636 | 0.18 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr19_-_47571797 | 0.18 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr17_+_14711765 | 0.18 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr12_+_5102670 | 0.17 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr1_-_6028876 | 0.17 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr15_+_1534644 | 0.17 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr23_+_2666944 | 0.17 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr16_+_23487051 | 0.17 |
ENSDART00000145496
|
icn2
|
ictacalcin 2 |
| chr21_-_39024754 | 0.17 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr24_-_39826865 | 0.17 |
ENSDART00000089232
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr15_-_1843831 | 0.17 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr12_-_46252062 | 0.17 |
ENSDART00000153223
|
si:ch211-226h7.5
|
si:ch211-226h7.5 |
| chr15_+_14856307 | 0.16 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr5_-_29531948 | 0.16 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr21_-_17296789 | 0.16 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr14_+_32918172 | 0.16 |
ENSDART00000182867
|
lnx2b
|
ligand of numb-protein X 2b |
| chr7_+_36041509 | 0.16 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr19_-_2317558 | 0.16 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr21_+_13327527 | 0.16 |
ENSDART00000114294
|
snrpd3l
|
small nuclear ribonucleoprotein D3 polypeptide, like |
| chr10_+_6013076 | 0.16 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr1_-_9249943 | 0.16 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr3_-_32957702 | 0.16 |
ENSDART00000146586
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr20_+_26880668 | 0.15 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr3_+_6469754 | 0.15 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr23_+_43718115 | 0.15 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr5_-_42883761 | 0.15 |
ENSDART00000167374
|
BX323596.2
|
|
| chr2_+_58841181 | 0.15 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr20_+_19175031 | 0.15 |
ENSDART00000138028
|
blk
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr16_-_46645396 | 0.15 |
ENSDART00000131485
|
tmem176l.2
|
transmembrane protein 176l.2 |
| chr24_+_18286427 | 0.15 |
ENSDART00000055443
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr20_+_52546186 | 0.15 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr9_+_41224100 | 0.15 |
ENSDART00000141792
|
stat1b
|
signal transducer and activator of transcription 1b |
| chr11_+_18130300 | 0.15 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr8_+_30709685 | 0.15 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr15_-_4580763 | 0.15 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr1_+_44173245 | 0.15 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr2_+_52847049 | 0.14 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr21_-_19918286 | 0.14 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr2_+_35595454 | 0.14 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr20_+_30610547 | 0.14 |
ENSDART00000122256
|
ccr6a
|
chemokine (C-C motif) receptor 6a |
| chr7_+_1579510 | 0.14 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr20_+_18702624 | 0.14 |
ENSDART00000185605
ENSDART00000019476 ENSDART00000152533 |
eif5
|
eukaryotic translation initiation factor 5 |
| chr19_-_18626515 | 0.14 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr14_+_48862987 | 0.14 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr8_+_3379815 | 0.14 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr15_-_563877 | 0.14 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr24_+_17007407 | 0.14 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr3_-_34089310 | 0.14 |
ENSDART00000151774
|
ighv5-5
|
immunoglobulin heavy variable 5-5 |
| chr10_-_22127942 | 0.14 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr9_+_20853894 | 0.14 |
ENSDART00000003648
|
wdr3
|
WD repeat domain 3 |
| chr2_-_26596794 | 0.14 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr10_+_32050906 | 0.14 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr24_+_36840652 | 0.13 |
ENSDART00000088168
|
CABZ01055347.1
|
|
| chr2_+_25839940 | 0.13 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr8_+_6576940 | 0.13 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr15_-_31027112 | 0.13 |
ENSDART00000100185
|
lgals9l4
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 4 |
| chr21_-_2415808 | 0.13 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr10_+_15454745 | 0.13 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr2_-_43151358 | 0.13 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr4_-_18954001 | 0.13 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr4_-_12723585 | 0.13 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr3_-_55104310 | 0.13 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr9_-_34269066 | 0.13 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr12_-_46112892 | 0.13 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr11_+_18157260 | 0.13 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr5_-_57723929 | 0.12 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr5_-_54712159 | 0.12 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr1_+_45056371 | 0.12 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr19_-_27550768 | 0.12 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr9_+_23770666 | 0.12 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr5_-_1274794 | 0.12 |
ENSDART00000167337
ENSDART00000125247 |
ciz1a
|
cdkn1a interacting zinc finger protein 1a |
| chr1_-_56176976 | 0.12 |
ENSDART00000052688
|
c3a.1
|
complement component c3a, duplicate 1 |
| chr6_+_3693441 | 0.12 |
ENSDART00000065256
|
ppig
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr13_+_36585399 | 0.12 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr18_+_31056645 | 0.12 |
ENSDART00000159316
|
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr19_-_617246 | 0.12 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr14_+_14841685 | 0.12 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr7_+_49681040 | 0.12 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr18_-_3520358 | 0.12 |
ENSDART00000181412
|
capn5a
|
calpain 5a |
| chr21_+_33459524 | 0.12 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr21_-_32781612 | 0.12 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr20_-_25522911 | 0.12 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr8_+_50190742 | 0.12 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr21_+_16980141 | 0.12 |
ENSDART00000101241
|
aqp3b
|
aquaporin 3b |
| chr8_-_12432604 | 0.12 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr2_-_59145027 | 0.12 |
ENSDART00000128320
|
FO834803.1
|
|
| chr11_+_14284866 | 0.11 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr6_-_54290227 | 0.11 |
ENSDART00000050483
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr20_+_29565906 | 0.11 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr17_+_39741926 | 0.11 |
ENSDART00000154996
ENSDART00000154599 |
si:dkey-229e3.2
|
si:dkey-229e3.2 |
| chr12_-_30540699 | 0.11 |
ENSDART00000167712
ENSDART00000102464 |
zgc:153920
|
zgc:153920 |
| chr3_-_26184018 | 0.11 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr10_-_10969596 | 0.11 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr19_-_30447611 | 0.11 |
ENSDART00000073705
ENSDART00000048977 ENSDART00000191237 |
abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr2_+_25839650 | 0.11 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr6_-_42388608 | 0.11 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr7_-_57637779 | 0.11 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr10_-_10969444 | 0.11 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr17_+_31221761 | 0.11 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr14_-_36862745 | 0.11 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr22_+_10440991 | 0.11 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr8_+_39724138 | 0.11 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr24_-_1985007 | 0.11 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr16_+_35916371 | 0.11 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr17_+_25331576 | 0.11 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr22_+_34616151 | 0.11 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr3_-_4501026 | 0.11 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr7_-_26457208 | 0.11 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr14_+_31496543 | 0.11 |
ENSDART00000170683
|
phf6
|
PHD finger protein 6 |
| chr10_-_1276046 | 0.11 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr25_-_21031007 | 0.11 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr2_-_10877765 | 0.10 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr22_+_6254194 | 0.10 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr25_+_35913614 | 0.10 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr7_-_16598212 | 0.10 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr16_-_30421275 | 0.10 |
ENSDART00000003752
|
cct3
|
chaperonin containing TCP1, subunit 3 (gamma) |
| chr10_-_35410518 | 0.10 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr11_+_575665 | 0.10 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr3_+_32139666 | 0.10 |
ENSDART00000109791
|
zgc:92066
|
zgc:92066 |
| chr25_+_3994823 | 0.10 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr11_+_16166117 | 0.10 |
ENSDART00000109585
ENSDART00000135724 |
hmces
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr2_+_36112273 | 0.10 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr5_+_66433287 | 0.10 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr6_-_9282080 | 0.10 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr3_+_32135037 | 0.10 |
ENSDART00000110490
|
BX511021.1
|
|
| chr21_-_14826066 | 0.10 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr8_+_3405612 | 0.10 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr7_+_73801377 | 0.10 |
ENSDART00000184051
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr8_+_23152244 | 0.10 |
ENSDART00000036801
ENSDART00000184298 |
slc17a9a
|
solute carrier family 17 (vesicular nucleotide transporter), member 9a |
| chr8_+_39674707 | 0.10 |
ENSDART00000126301
ENSDART00000040330 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr1_-_53468160 | 0.10 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr18_+_29898955 | 0.10 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr7_-_18508815 | 0.10 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr20_-_49889111 | 0.10 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr12_+_3723650 | 0.10 |
ENSDART00000179922
ENSDART00000152482 ENSDART00000108771 |
pagr1
|
PAXIP1 associated glutamate-rich protein 1 |
| chr7_+_24522308 | 0.10 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr22_-_10440688 | 0.10 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr8_+_45334255 | 0.10 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr21_-_3007412 | 0.09 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr5_-_38107741 | 0.09 |
ENSDART00000156853
|
si:ch211-284e13.14
|
si:ch211-284e13.14 |
| chr17_+_45817527 | 0.09 |
ENSDART00000035152
|
kif26ab
|
kinesin family member 26Ab |
| chr17_-_31611692 | 0.09 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr17_+_27456804 | 0.09 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr20_-_34670236 | 0.09 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr7_+_2228276 | 0.09 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr5_-_33281046 | 0.09 |
ENSDART00000051344
ENSDART00000138116 |
surf6
|
surfeit 6 |
| chr3_+_49097775 | 0.09 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr3_+_32105175 | 0.09 |
ENSDART00000113789
|
zgc:198419
|
zgc:198419 |
| chr6_-_18228358 | 0.09 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr1_-_13989643 | 0.09 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr18_+_6558338 | 0.09 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr12_-_20616160 | 0.09 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr13_-_14908999 | 0.09 |
ENSDART00000112771
|
ap5s1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr3_-_15999501 | 0.09 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr23_+_10146542 | 0.09 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr16_-_32304764 | 0.09 |
ENSDART00000143859
ENSDART00000134381 |
mms22l
|
MMS22-like, DNA repair protein |
| chr6_+_45918981 | 0.09 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr23_+_11285662 | 0.09 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr22_-_11661724 | 0.09 |
ENSDART00000025198
|
mettl21a
|
methyltransferase like 21A |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.2 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.2 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:1902230 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.0 | 0.3 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.1 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.0 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.0 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.0 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.0 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.0 | 0.1 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.0 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.1 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.1 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.0 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.0 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |