Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
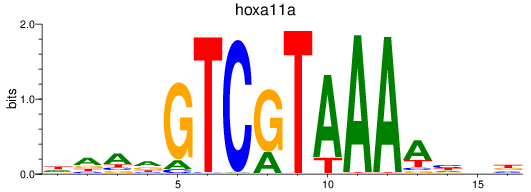
Results for hoxa11a
Z-value: 0.32
Transcription factors associated with hoxa11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11a
|
ENSDARG00000104162 | homeobox A11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11a | dr11_v1_chr19_+_19737214_19737300 | -0.69 | 2.0e-01 | Click! |
Activity profile of hoxa11a motif
Sorted Z-values of hoxa11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_9586713 | 0.21 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr7_+_31879649 | 0.21 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr8_-_14049404 | 0.21 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr7_+_31879986 | 0.19 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr10_+_33990572 | 0.17 |
ENSDART00000138547
|
fryb
|
furry homolog b (Drosophila) |
| chr6_-_40744720 | 0.14 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr7_+_20524064 | 0.13 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr7_+_20966434 | 0.13 |
ENSDART00000185570
|
efnb3b
|
ephrin-B3b |
| chr18_+_18000887 | 0.13 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr12_+_33361948 | 0.10 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr14_-_31060082 | 0.10 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr22_+_10646928 | 0.09 |
ENSDART00000038465
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr16_-_6205790 | 0.08 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr5_+_36611128 | 0.08 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr4_-_20108833 | 0.08 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr12_-_26851726 | 0.08 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr24_+_21540842 | 0.08 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr6_-_53334259 | 0.08 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr3_-_27868183 | 0.08 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr24_-_40009446 | 0.07 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr20_-_44576949 | 0.07 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr25_-_18002937 | 0.07 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr9_-_3671911 | 0.07 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr5_-_35301800 | 0.07 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr11_-_26832685 | 0.07 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr22_-_10502780 | 0.07 |
ENSDART00000136961
|
ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr14_-_48348973 | 0.06 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr2_-_13254821 | 0.06 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr9_-_23922011 | 0.06 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr5_+_59397348 | 0.06 |
ENSDART00000175642
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr18_+_40993369 | 0.06 |
ENSDART00000141162
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr5_-_32489796 | 0.06 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr3_+_36284986 | 0.05 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr17_-_23727978 | 0.05 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr25_-_18002498 | 0.05 |
ENSDART00000158688
|
cep290
|
centrosomal protein 290 |
| chr3_+_24190207 | 0.05 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr4_+_39368978 | 0.05 |
ENSDART00000160640
|
si:dkey-261o4.1
|
si:dkey-261o4.1 |
| chr15_+_36115955 | 0.05 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr5_+_59397739 | 0.05 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr2_-_13254594 | 0.05 |
ENSDART00000155671
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr16_-_24561354 | 0.05 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr15_+_8767650 | 0.04 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr18_-_2433011 | 0.04 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr2_-_43851915 | 0.04 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr11_+_43751263 | 0.04 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr22_+_2239974 | 0.04 |
ENSDART00000141993
|
znf1144
|
zinc finger protein 1144 |
| chr7_-_43840418 | 0.04 |
ENSDART00000174592
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr2_-_43852207 | 0.04 |
ENSDART00000192627
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr12_-_34713690 | 0.03 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr15_+_44093286 | 0.03 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr7_-_18508815 | 0.03 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr13_-_9875538 | 0.03 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr1_+_36722122 | 0.03 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr3_-_40254634 | 0.03 |
ENSDART00000154562
|
top3a
|
DNA topoisomerase III alpha |
| chr10_-_25561751 | 0.03 |
ENSDART00000147089
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr7_-_30280934 | 0.03 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr22_-_33916620 | 0.03 |
ENSDART00000191276
|
CR974456.1
|
|
| chr7_-_41014773 | 0.03 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr2_-_9259283 | 0.03 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr6_+_33885828 | 0.02 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr6_+_49551614 | 0.02 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr5_-_31856681 | 0.02 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr25_-_19395156 | 0.02 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr24_-_31223232 | 0.02 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr13_+_35472803 | 0.02 |
ENSDART00000011583
|
mkks
|
McKusick-Kaufman syndrome |
| chr24_+_20658760 | 0.02 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr16_-_47426482 | 0.02 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr11_+_25560632 | 0.01 |
ENSDART00000033914
|
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr18_+_40993196 | 0.01 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr25_+_3788443 | 0.01 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr24_+_20658942 | 0.01 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr17_+_4368859 | 0.01 |
ENSDART00000055385
|
crls1
|
cardiolipin synthase 1 |
| chr12_-_11560794 | 0.01 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr7_-_30624435 | 0.01 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr11_+_25560072 | 0.01 |
ENSDART00000124131
ENSDART00000147179 |
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr16_-_10316359 | 0.01 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr9_-_29039506 | 0.01 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr13_+_45431660 | 0.01 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr11_-_21303946 | 0.01 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr13_+_29925397 | 0.00 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr15_-_14552101 | 0.00 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_+_44463230 | 0.00 |
ENSDART00000130311
|
naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr6_+_3730843 | 0.00 |
ENSDART00000019630
|
FO704755.1
|
|
| chr18_-_17087138 | 0.00 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.2 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |