Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
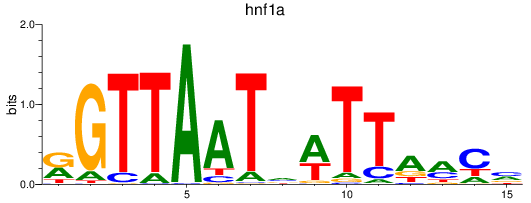
Results for hnf1a
Z-value: 4.92
Transcription factors associated with hnf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf1a
|
ENSDARG00000009470 | HNF1 homeobox a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf1a | dr11_v1_chr8_+_39802506_39802506 | 0.85 | 6.9e-02 | Click! |
Activity profile of hnf1a motif
Sorted Z-values of hnf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24757785 | 7.09 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr22_-_24791505 | 7.06 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr22_-_23625351 | 6.87 |
ENSDART00000192588
ENSDART00000163423 |
cfhl4
|
complement factor H like 4 |
| chr22_-_24858042 | 6.74 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr1_+_10051763 | 6.69 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr22_-_24738188 | 6.04 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr13_+_23988442 | 6.03 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr5_+_32345187 | 5.52 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr5_+_45677781 | 5.42 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr23_+_25856541 | 5.34 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr9_-_9982696 | 5.26 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr21_+_40092301 | 5.20 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr22_-_24818066 | 5.00 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr22_-_23668356 | 4.95 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr21_-_45920 | 4.90 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr20_-_40750953 | 4.77 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr2_-_5475910 | 4.70 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr11_-_37997419 | 4.56 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr5_+_28830643 | 4.50 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr12_-_6172154 | 4.43 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr12_-_6177894 | 4.36 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr1_-_12126535 | 4.32 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr5_+_28830388 | 4.18 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr22_+_12770877 | 4.14 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr15_-_33965440 | 4.13 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_-_4296771 | 4.09 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr5_+_28849155 | 4.07 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr5_+_28848870 | 4.06 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr13_-_40726865 | 4.03 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr24_-_17039638 | 3.97 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr1_-_59348118 | 3.84 |
ENSDART00000170901
|
cyp3a65
|
cytochrome P450, family 3, subfamily A, polypeptide 65 |
| chr15_-_33964897 | 3.80 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr20_+_10544100 | 3.80 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr2_-_41942666 | 3.77 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr22_-_23706771 | 3.76 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr9_-_35633827 | 3.75 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr10_+_21867307 | 3.68 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr17_+_4030493 | 3.62 |
ENSDART00000151849
|
hao1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr22_-_23748284 | 3.43 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr4_+_18804317 | 3.39 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr2_-_5466708 | 3.35 |
ENSDART00000136682
|
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr9_-_34269066 | 3.35 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr5_+_28858345 | 3.20 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr18_+_15644559 | 3.14 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr2_-_127945 | 3.14 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr3_+_49521106 | 3.13 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr2_+_26647472 | 3.06 |
ENSDART00000145415
ENSDART00000157409 |
ttpa
|
tocopherol (alpha) transfer protein |
| chr8_-_2616326 | 3.01 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr5_+_26795773 | 2.93 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr3_+_15505275 | 2.79 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr6_-_59563597 | 2.78 |
ENSDART00000166311
|
INHBE
|
inhibin beta E subunit |
| chr22_-_23781083 | 2.69 |
ENSDART00000166563
ENSDART00000170458 ENSDART00000166158 ENSDART00000171246 |
cfhl3
|
complement factor H like 3 |
| chr15_+_9297340 | 2.60 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr25_+_31323978 | 2.52 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr16_-_17200120 | 2.38 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr13_+_2394264 | 2.29 |
ENSDART00000168595
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr5_-_30620625 | 2.17 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr11_-_23332592 | 2.16 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr6_+_8622228 | 2.16 |
ENSDART00000110470
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr8_-_13013123 | 2.15 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr12_-_20373058 | 2.14 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr22_-_17611742 | 2.11 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr2_+_27855102 | 2.10 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr5_+_9405283 | 2.08 |
ENSDART00000127306
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr3_+_26734162 | 2.06 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr22_-_17595310 | 2.05 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr12_+_2648043 | 2.04 |
ENSDART00000082220
|
gdf2
|
growth differentiation factor 2 |
| chr7_-_10560964 | 2.03 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr5_+_9382301 | 2.03 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr23_+_26491931 | 2.00 |
ENSDART00000190124
ENSDART00000132905 |
si:ch73-206d17.1
|
si:ch73-206d17.1 |
| chr1_-_9134045 | 1.97 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr21_+_38002879 | 1.96 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr8_+_999421 | 1.94 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr9_+_21151138 | 1.90 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr8_-_40555340 | 1.88 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr2_-_24402341 | 1.85 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr7_-_35066457 | 1.82 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr25_-_27541621 | 1.80 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr13_-_42579437 | 1.73 |
ENSDART00000135172
|
fam83ha
|
family with sequence similarity 83, member Ha |
| chr5_+_57480014 | 1.73 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr14_-_17622080 | 1.69 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr6_+_7977611 | 1.67 |
ENSDART00000112290
|
palm3
|
paralemmin 3 |
| chr5_-_37886063 | 1.66 |
ENSDART00000131378
ENSDART00000132152 |
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr7_-_61901695 | 1.64 |
ENSDART00000110328
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr5_+_9417409 | 1.63 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr5_+_9377005 | 1.57 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr24_-_4765740 | 1.57 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr25_-_27541288 | 1.53 |
ENSDART00000187245
|
spam1
|
sperm adhesion molecule 1 |
| chr12_+_13091842 | 1.52 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr7_+_40205394 | 1.51 |
ENSDART00000173742
ENSDART00000148390 |
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr10_-_34915886 | 1.51 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr11_-_27962757 | 1.44 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr2_-_43635777 | 1.42 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr25_+_29474583 | 1.37 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr6_+_41039166 | 1.34 |
ENSDART00000125659
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr23_-_31060350 | 1.33 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr3_+_12718100 | 1.33 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr7_+_2849020 | 1.30 |
ENSDART00000168695
|
BX004857.1
|
|
| chr21_+_25236297 | 1.29 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr13_-_23666579 | 1.28 |
ENSDART00000192457
ENSDART00000179777 |
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr6_-_39344259 | 1.27 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr11_+_43419809 | 1.25 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr14_-_32884138 | 1.23 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr7_-_29534001 | 1.22 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr10_-_34916208 | 1.21 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr17_+_30591287 | 1.21 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr16_-_42894628 | 1.21 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr2_-_58183499 | 1.20 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr20_+_6543674 | 1.20 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr21_-_31286428 | 1.18 |
ENSDART00000185334
|
ca4c
|
carbonic anhydrase IV c |
| chr14_-_11507211 | 1.16 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr25_+_29474982 | 1.15 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr20_+_35473288 | 1.14 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr17_-_34952562 | 1.08 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr24_-_23675446 | 1.08 |
ENSDART00000066644
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr25_+_34915762 | 1.05 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr3_+_25191467 | 1.05 |
ENSDART00000156956
ENSDART00000154799 |
il2rb
|
interleukin 2 receptor, beta |
| chr12_+_5209822 | 1.04 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr20_+_48754045 | 1.03 |
ENSDART00000062578
|
zgc:110348
|
zgc:110348 |
| chr24_-_20016817 | 1.03 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr2_+_17055069 | 1.02 |
ENSDART00000115078
|
thpo
|
thrombopoietin |
| chr14_-_41556720 | 1.02 |
ENSDART00000149244
|
itga6l
|
integrin, alpha 6, like |
| chr25_+_23280220 | 1.02 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr24_-_34680956 | 1.02 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr2_-_36500347 | 0.99 |
ENSDART00000110528
|
BX901889.1
|
|
| chr15_-_23467750 | 0.99 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr9_+_29994439 | 0.97 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr21_-_22647695 | 0.97 |
ENSDART00000187553
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr3_+_31039923 | 0.96 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr3_-_6191950 | 0.95 |
ENSDART00000161931
|
si:ch73-144l3.1
|
si:ch73-144l3.1 |
| chr4_+_57881965 | 0.93 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr18_+_15683157 | 0.91 |
ENSDART00000140933
|
si:ch211-264e16.2
|
si:ch211-264e16.2 |
| chr3_+_49097775 | 0.91 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr24_+_5811808 | 0.91 |
ENSDART00000132428
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr20_+_18740518 | 0.91 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr6_-_43498996 | 0.87 |
ENSDART00000154742
|
MDFIC2
|
si:dkey-58f6.2 |
| chr22_+_15960514 | 0.87 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr3_+_13559199 | 0.86 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr6_-_43283122 | 0.84 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr6_+_34028532 | 0.83 |
ENSDART00000155827
|
si:ch73-185c24.2
|
si:ch73-185c24.2 |
| chr25_-_7670616 | 0.80 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr5_+_1877464 | 0.80 |
ENSDART00000050658
|
zgc:101699
|
zgc:101699 |
| chr22_+_15960005 | 0.80 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr19_+_28256076 | 0.79 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr6_-_37744430 | 0.77 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr5_-_67783593 | 0.75 |
ENSDART00000010934
|
casr
|
calcium-sensing receptor |
| chr17_+_15388479 | 0.73 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr18_+_13182528 | 0.71 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr9_+_30720048 | 0.71 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr16_+_55059026 | 0.71 |
ENSDART00000109391
|
LO017815.1
|
Danio rerio nuclear receptor coactivator 7-like (LOC792958), mRNA. |
| chr16_+_11623956 | 0.71 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr15_+_14109652 | 0.70 |
ENSDART00000156370
|
clca1
|
chloride channel accessory 1 |
| chr3_-_53092509 | 0.70 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr8_+_1187928 | 0.70 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr5_-_63302944 | 0.69 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr16_+_11603096 | 0.69 |
ENSDART00000146286
|
si:dkey-11o1.3
|
si:dkey-11o1.3 |
| chr20_+_18740799 | 0.67 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr25_+_34915576 | 0.66 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr12_+_15666949 | 0.66 |
ENSDART00000079803
|
nmt1b
|
N-myristoyltransferase 1b |
| chr3_-_53091946 | 0.66 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr11_-_12800945 | 0.64 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr17_+_1496107 | 0.63 |
ENSDART00000187804
|
LO018430.1
|
|
| chr3_-_18047150 | 0.63 |
ENSDART00000020376
|
hsd17b1
|
hydroxysteroid (17-beta) dehydrogenase 1 |
| chr4_-_56954002 | 0.61 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr11_-_19501439 | 0.61 |
ENSDART00000158328
ENSDART00000103983 |
slc2a9l1
|
solute carrier family 2 (facilitated glucose transporter), member 9-like 1 |
| chr6_+_40723554 | 0.61 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr14_+_46118834 | 0.61 |
ENSDART00000124417
ENSDART00000017785 |
naa15a
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit a |
| chr13_+_18523661 | 0.61 |
ENSDART00000034852
|
tlr4bb
|
toll-like receptor 4b, duplicate b |
| chr11_-_12801157 | 0.60 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr21_+_11535304 | 0.59 |
ENSDART00000158901
ENSDART00000081600 |
cd8b
|
cd8 beta |
| chr13_+_24853578 | 0.58 |
ENSDART00000145865
|
si:dkey-24f15.2
|
si:dkey-24f15.2 |
| chr20_-_25481306 | 0.58 |
ENSDART00000182495
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr7_-_25126212 | 0.54 |
ENSDART00000173588
|
MAJIN
|
si:dkey-23i12.11 |
| chr2_-_10821053 | 0.54 |
ENSDART00000056034
|
rpap2
|
RNA polymerase II associated protein 2 |
| chr14_+_31721171 | 0.53 |
ENSDART00000173034
|
adgrg4a
|
adhesion G protein-coupled receptor G4a |
| chr23_-_20345473 | 0.51 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr2_+_36121373 | 0.51 |
ENSDART00000187002
|
CT867973.2
|
|
| chr3_-_16250527 | 0.51 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr16_-_31980948 | 0.50 |
ENSDART00000182427
|
CR854927.4
|
|
| chr20_-_43723860 | 0.49 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr22_+_15959844 | 0.48 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr6_+_30456788 | 0.47 |
ENSDART00000121492
|
FP236735.1
|
|
| chr25_+_16194979 | 0.47 |
ENSDART00000185592
ENSDART00000158582 ENSDART00000161109 ENSDART00000139013 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr19_-_15229421 | 0.46 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr2_-_23778180 | 0.46 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr11_-_40647190 | 0.45 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr25_-_37191929 | 0.44 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr12_+_6117682 | 0.43 |
ENSDART00000146650
|
asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr7_+_67429185 | 0.43 |
ENSDART00000162553
ENSDART00000178646 |
kars
|
lysyl-tRNA synthetase |
| chr6_-_32087108 | 0.42 |
ENSDART00000150934
ENSDART00000130627 |
alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr19_+_31043002 | 0.42 |
ENSDART00000171591
|
ankmy2b
|
ankyrin repeat and MYND domain containing 2b |
| chr16_+_11724230 | 0.41 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr8_+_17869225 | 0.40 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr14_-_41556533 | 0.40 |
ENSDART00000074401
|
itga6l
|
integrin, alpha 6, like |
| chr16_-_29403711 | 0.38 |
ENSDART00000175571
|
tlr18
|
toll-like receptor 18 |
| chr4_-_14470071 | 0.37 |
ENSDART00000143773
|
plxnb2a
|
plexin b2a |
| chr5_+_45322734 | 0.37 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr23_+_33752275 | 0.37 |
ENSDART00000007260
|
si:ch211-210c8.6
|
si:ch211-210c8.6 |
| chr15_+_27364394 | 0.34 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr4_-_12766418 | 0.34 |
ENSDART00000024312
|
dera
|
deoxyribose-phosphate aldolase (putative) |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.4 | 4.1 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 1.3 | 3.8 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 1.0 | 2.9 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.8 | 32.2 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.6 | 8.8 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.5 | 4.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.5 | 5.3 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.4 | 6.7 | GO:0070527 | platelet aggregation(GO:0070527) blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 4.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.4 | 35.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.4 | 2.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 2.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.3 | 8.5 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 4.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.3 | 1.2 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.3 | 2.0 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.3 | 3.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 1.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 2.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 2.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 0.7 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.2 | 1.3 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 3.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 3.3 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.2 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 3.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.5 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.2 | 8.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 3.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.7 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 2.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.6 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.3 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 1.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 8.4 | GO:0007492 | endoderm development(GO:0007492) |
| 0.1 | 1.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 3.8 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 2.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.2 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 1.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 2.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 5.5 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 1.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 2.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.0 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.6 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.3 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.0 | 0.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 3.4 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 1.1 | GO:0001894 | tissue homeostasis(GO:0001894) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.8 | 5.5 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.9 | 2.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 2.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 2.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.9 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 1.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 4.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 52.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 3.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 31.9 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 1.8 | 5.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.3 | 4.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.1 | 4.3 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 1.0 | 4.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.8 | 4.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.6 | 3.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.6 | 2.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.5 | 4.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.4 | 2.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 1.0 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.3 | 1.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.3 | 35.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 3.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.4 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 5.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 2.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 2.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 4.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 0.7 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 12.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 2.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 0.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 0.7 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.2 | 0.6 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 8.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.9 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 2.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 1.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.5 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 4.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 5.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 3.7 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 9.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 2.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 5.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.0 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.2 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 2.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 6.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 3.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 2.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 11.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.6 | 6.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 4.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 9.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 2.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 6.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 2.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 1.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 6.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 2.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 6.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 7.3 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |