Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
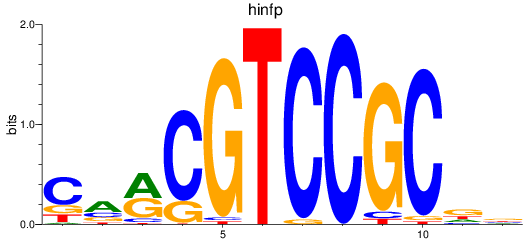
Results for hinfp
Z-value: 0.81
Transcription factors associated with hinfp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hinfp
|
ENSDARG00000004851 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hinfp | dr11_v1_chr5_+_30596477_30596477 | 0.58 | 3.0e-01 | Click! |
Activity profile of hinfp motif
Sorted Z-values of hinfp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_62244744 | 0.27 |
ENSDART00000192522
ENSDART00000170635 ENSDART00000073872 |
CCKAR
|
cholecystokinin A receptor |
| chr3_+_6469754 | 0.25 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr13_-_3155243 | 0.24 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr1_+_53945934 | 0.23 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr19_-_11425542 | 0.21 |
ENSDART00000177875
ENSDART00000080762 |
sept7a
|
septin 7a |
| chr17_+_6793001 | 0.20 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr5_+_64407896 | 0.20 |
ENSDART00000173886
|
si:ch1073-363c19.3
|
si:ch1073-363c19.3 |
| chr8_+_40644838 | 0.19 |
ENSDART00000169311
|
adra2b
|
adrenoceptor alpha 2B |
| chr5_-_37881345 | 0.16 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr2_+_15048410 | 0.15 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr21_-_41147818 | 0.14 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr21_-_12749501 | 0.14 |
ENSDART00000179724
|
LO018011.1
|
|
| chr11_+_6010177 | 0.14 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr5_-_63065912 | 0.13 |
ENSDART00000161149
|
fam57a
|
family with sequence similarity 57 member A |
| chr11_-_21363834 | 0.13 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr23_+_642001 | 0.13 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr5_-_23117078 | 0.12 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr13_+_52006253 | 0.12 |
ENSDART00000181259
|
CABZ01080135.1
|
|
| chr6_+_7553085 | 0.12 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr22_+_11775269 | 0.12 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr5_-_47975440 | 0.12 |
ENSDART00000145665
ENSDART00000007057 |
ccnh
|
cyclin H |
| chr10_-_35321625 | 0.12 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr7_-_48396193 | 0.12 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr22_+_38310957 | 0.11 |
ENSDART00000040550
|
traf5
|
Tnf receptor-associated factor 5 |
| chr8_-_44904723 | 0.11 |
ENSDART00000040804
|
praf2
|
PRA1 domain family, member 2 |
| chr1_+_54069450 | 0.11 |
ENSDART00000108601
ENSDART00000187878 |
dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr9_+_30890549 | 0.11 |
ENSDART00000101070
|
dachd
|
dachshund d |
| chr19_-_1023051 | 0.11 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr1_-_40132831 | 0.10 |
ENSDART00000135647
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr4_-_20081621 | 0.10 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr7_-_29231686 | 0.10 |
ENSDART00000173780
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr19_+_58954 | 0.10 |
ENSDART00000162379
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr2_-_1468258 | 0.09 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr11_+_6009984 | 0.09 |
ENSDART00000185680
|
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr6_-_40771813 | 0.08 |
ENSDART00000076200
|
h1fx
|
H1 histone family, member X |
| chr20_-_2725594 | 0.08 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr23_+_30707837 | 0.08 |
ENSDART00000016096
|
dnajc11a
|
DnaJ (Hsp40) homolog, subfamily C, member 11a |
| chr8_+_11687586 | 0.08 |
ENSDART00000146241
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr22_-_20342260 | 0.08 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr7_+_63325819 | 0.08 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr8_+_21225064 | 0.08 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr15_-_25319663 | 0.08 |
ENSDART00000042218
|
pafah1b1a
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1a |
| chr21_+_28747236 | 0.08 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr21_+_28747069 | 0.08 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr22_+_31059919 | 0.07 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr2_-_27651674 | 0.07 |
ENSDART00000177402
|
tgs1
|
trimethylguanosine synthase 1 |
| chr21_-_41839683 | 0.07 |
ENSDART00000160908
ENSDART00000137630 |
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr3_+_26035550 | 0.07 |
ENSDART00000078137
|
ankrd54
|
ankyrin repeat domain 54 |
| chr12_+_46608361 | 0.06 |
ENSDART00000189164
|
FADS6
|
fatty acid desaturase 6 |
| chr3_-_21185016 | 0.06 |
ENSDART00000155550
|
taok2a
|
TAO kinase 2a |
| chr16_+_9726038 | 0.06 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr11_-_25853212 | 0.06 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr5_+_47863153 | 0.06 |
ENSDART00000051518
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr5_-_18961694 | 0.06 |
ENSDART00000142531
ENSDART00000090521 |
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr20_-_43917647 | 0.06 |
ENSDART00000026213
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr16_+_52771199 | 0.06 |
ENSDART00000111383
|
baalca
|
brain and acute leukemia, cytoplasmic a |
| chr16_+_25806400 | 0.06 |
ENSDART00000154652
|
irgq1
|
immunity-related GTPase family, q1 |
| chr8_+_11687254 | 0.05 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr20_-_2725930 | 0.05 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr20_-_2298970 | 0.05 |
ENSDART00000136067
|
EPB41L2
|
si:ch73-18b11.1 |
| chr3_-_6441619 | 0.05 |
ENSDART00000157771
ENSDART00000166758 |
sumo2b
|
small ubiquitin-like modifier 2b |
| chr10_+_13279079 | 0.05 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr16_+_2905150 | 0.05 |
ENSDART00000109980
|
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr23_-_44848961 | 0.05 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr19_+_48290786 | 0.05 |
ENSDART00000165986
|
zgc:101785
|
zgc:101785 |
| chr19_+_5135113 | 0.05 |
ENSDART00000151310
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr8_+_36983559 | 0.05 |
ENSDART00000127053
|
kdm5c
|
lysine (K)-specific demethylase 5C |
| chr21_+_21279159 | 0.05 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr21_+_35215810 | 0.05 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr1_+_40801448 | 0.05 |
ENSDART00000187394
|
cpz
|
carboxypeptidase Z |
| chr13_+_29925397 | 0.04 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr14_+_14992909 | 0.04 |
ENSDART00000182203
|
BX119314.1
|
|
| chr9_+_56422311 | 0.04 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr20_+_42246948 | 0.04 |
ENSDART00000061135
|
gopc
|
golgi-associated PDZ and coiled-coil motif containing |
| chr17_-_24866964 | 0.04 |
ENSDART00000190601
ENSDART00000192547 |
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr8_-_37461835 | 0.04 |
ENSDART00000013446
|
eif2d
|
eukaryotic translation initiation factor 2D |
| chr8_+_29962635 | 0.04 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr7_+_29163762 | 0.04 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr17_+_5846202 | 0.04 |
ENSDART00000189713
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr4_-_77557279 | 0.04 |
ENSDART00000180113
|
AL935186.10
|
|
| chr10_+_21444654 | 0.04 |
ENSDART00000140113
ENSDART00000184386 ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr25_-_10791437 | 0.04 |
ENSDART00000127054
|
BX572619.1
|
|
| chr7_+_28612671 | 0.04 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr10_-_39130839 | 0.04 |
ENSDART00000061274
ENSDART00000148648 |
rps25
|
ribosomal protein S25 |
| chr19_-_31900893 | 0.04 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr5_+_31168096 | 0.04 |
ENSDART00000086443
ENSDART00000192271 |
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr25_-_31907590 | 0.04 |
ENSDART00000149707
ENSDART00000149471 |
eif3ja
|
eukaryotic translation initiation factor 3, subunit Ja |
| chr11_+_1764203 | 0.04 |
ENSDART00000173023
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr5_+_27429872 | 0.04 |
ENSDART00000087894
|
zgc:165555
|
zgc:165555 |
| chr5_+_47862636 | 0.04 |
ENSDART00000139824
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr6_-_57655299 | 0.03 |
ENSDART00000083807
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr10_+_21722892 | 0.03 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr4_+_27018663 | 0.03 |
ENSDART00000180778
|
CR749777.1
|
|
| chr23_-_45319592 | 0.03 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr21_+_18950575 | 0.03 |
ENSDART00000036172
|
crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr9_+_55154414 | 0.03 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr7_+_9880811 | 0.03 |
ENSDART00000128376
|
lins1
|
lines homolog 1 |
| chr7_-_69184420 | 0.03 |
ENSDART00000168311
ENSDART00000159239 ENSDART00000161319 |
usp10
|
ubiquitin specific peptidase 10 |
| chr4_+_11690923 | 0.03 |
ENSDART00000150624
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr4_-_6567355 | 0.03 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr8_+_49936585 | 0.03 |
ENSDART00000098707
|
naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr6_-_35310224 | 0.03 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr24_-_1023112 | 0.02 |
ENSDART00000147508
ENSDART00000066863 |
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr17_-_21784152 | 0.02 |
ENSDART00000127254
|
hmx2
|
H6 family homeobox 2 |
| chr6_-_57655030 | 0.02 |
ENSDART00000155438
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr9_-_4856767 | 0.02 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr16_-_17586883 | 0.02 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr5_-_8483457 | 0.02 |
ENSDART00000171886
ENSDART00000159477 ENSDART00000157423 |
lifra
|
leukemia inhibitory factor receptor alpha a |
| chr17_-_24866727 | 0.02 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr4_+_71086017 | 0.02 |
ENSDART00000159113
|
si:dkeyp-80d11.10
|
si:dkeyp-80d11.10 |
| chr15_-_419597 | 0.02 |
ENSDART00000058993
|
psph
|
phosphoserine phosphatase |
| chr11_+_2710530 | 0.02 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr9_+_56422517 | 0.02 |
ENSDART00000168620
|
gpr39
|
G protein-coupled receptor 39 |
| chr16_-_27564256 | 0.02 |
ENSDART00000078297
|
zgc:153215
|
zgc:153215 |
| chr25_-_26833100 | 0.02 |
ENSDART00000014052
|
neil1
|
nei-like DNA glycosylase 1 |
| chr25_+_37268900 | 0.01 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr23_+_1216215 | 0.01 |
ENSDART00000165957
|
utrn
|
utrophin |
| chr24_-_37568359 | 0.01 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr23_+_43668756 | 0.01 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr10_-_43928937 | 0.01 |
ENSDART00000168014
ENSDART00000086220 |
mctp1b
|
multiple C2 domains, transmembrane 1b |
| chr15_+_23550890 | 0.01 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr5_-_56943064 | 0.01 |
ENSDART00000146991
|
si:ch211-127d4.3
|
si:ch211-127d4.3 |
| chr16_+_27564270 | 0.01 |
ENSDART00000140460
|
tmem67
|
transmembrane protein 67 |
| chr7_-_39611109 | 0.01 |
ENSDART00000149991
|
ptprja
|
protein tyrosine phosphatase, receptor type, J a |
| chr11_+_5926850 | 0.01 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr10_+_8551897 | 0.01 |
ENSDART00000084353
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr16_+_23495600 | 0.01 |
ENSDART00000021092
|
snx27b
|
sorting nexin family member 27b |
| chr23_+_45845423 | 0.00 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr14_+_52440161 | 0.00 |
ENSDART00000168437
|
b4galt1l
|
DP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1, like |
| chr19_+_3653976 | 0.00 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr4_-_2196798 | 0.00 |
ENSDART00000110178
ENSDART00000149330 |
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr10_+_37981471 | 0.00 |
ENSDART00000113112
ENSDART00000185102 |
wscd1b
|
WSC domain containing 1b |
| chr6_+_34038963 | 0.00 |
ENSDART00000057732
ENSDART00000192496 |
ap1m3
|
adaptor-related protein complex 1, mu 3 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of hinfp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |