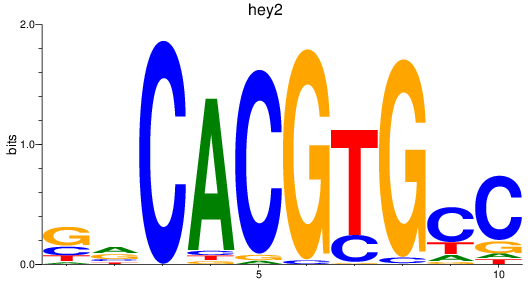
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
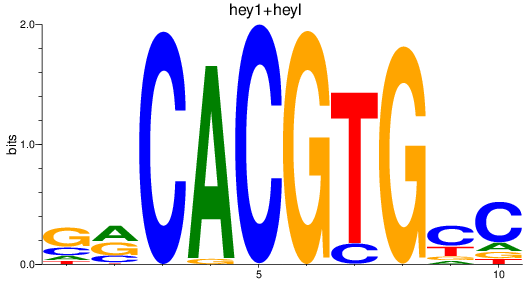
Results for hey2_hey1+heyl
Z-value: 1.34
Transcription factors associated with hey2_hey1+heyl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hey2
|
ENSDARG00000013441 | hes-related family bHLH transcription factor with YRPW motif 2 |
|
heyl
|
ENSDARG00000055798 | hes related family bHLH transcription factor with YRPW motif like |
|
hey1
|
ENSDARG00000070538 | hes-related family bHLH transcription factor with YRPW motif 1 |
|
heyl
|
ENSDARG00000112770 | hes related family bHLH transcription factor with YRPW motif like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| heyl | dr11_v1_chr19_+_32553874_32553874 | -0.31 | 6.1e-01 | Click! |
| hey1 | dr11_v1_chr19_-_31802296_31802296 | 0.23 | 7.1e-01 | Click! |
| hey2 | dr11_v1_chr20_-_39596338_39596338 | -0.16 | 7.9e-01 | Click! |
Activity profile of hey2_hey1+heyl motif
Sorted Z-values of hey2_hey1+heyl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_6780340 | 1.15 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr6_-_60147517 | 1.11 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr20_+_34915945 | 1.01 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr14_+_35748385 | 0.87 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr14_+_35748206 | 0.86 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr10_-_7913591 | 0.78 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr20_+_23440632 | 0.73 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr25_+_3328487 | 0.71 |
ENSDART00000181143
|
ldhbb
|
lactate dehydrogenase Bb |
| chr3_+_23488652 | 0.70 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr8_+_21146262 | 0.68 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr7_-_40993456 | 0.67 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr1_-_39943596 | 0.67 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr10_+_10677697 | 0.64 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr11_-_3552067 | 0.62 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr14_-_30387894 | 0.59 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr23_-_24856025 | 0.59 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr2_+_21090317 | 0.57 |
ENSDART00000109568
ENSDART00000139633 |
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr19_-_24555935 | 0.57 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr1_+_7956030 | 0.57 |
ENSDART00000159655
|
CR855320.1
|
|
| chr8_-_42238543 | 0.55 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr15_+_16897554 | 0.55 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr7_+_38717624 | 0.55 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr2_+_18988407 | 0.55 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr19_-_8940068 | 0.54 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr4_-_16824231 | 0.54 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr7_-_30082931 | 0.54 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr21_+_31150773 | 0.53 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr10_+_17026870 | 0.53 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr18_-_19103929 | 0.52 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr17_+_24318753 | 0.52 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr7_+_38750871 | 0.52 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr3_+_1015867 | 0.51 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr21_+_31150438 | 0.50 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr21_-_42100471 | 0.48 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_-_16824556 | 0.48 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr25_+_37268900 | 0.47 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr8_+_17184602 | 0.47 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr24_+_39135419 | 0.47 |
ENSDART00000180941
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr8_-_23081511 | 0.47 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr5_+_26075230 | 0.47 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr18_-_21218851 | 0.46 |
ENSDART00000060160
|
calb2a
|
calbindin 2a |
| chr17_-_17948587 | 0.46 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr18_+_19990412 | 0.45 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr18_-_40684756 | 0.45 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr13_-_4992395 | 0.45 |
ENSDART00000102651
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr2_+_20331445 | 0.44 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr2_+_29976419 | 0.44 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr24_-_38644937 | 0.44 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr5_+_43470544 | 0.44 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr11_-_3959889 | 0.43 |
ENSDART00000159683
|
pbrm1
|
polybromo 1 |
| chr19_-_24555623 | 0.43 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr1_+_17695426 | 0.43 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr22_+_438714 | 0.43 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr8_-_31606514 | 0.42 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr18_+_6479963 | 0.40 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr2_-_9489611 | 0.40 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr23_-_21463788 | 0.39 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr13_+_31402067 | 0.39 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr3_-_62380146 | 0.39 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr24_-_12938922 | 0.39 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr2_-_32513538 | 0.39 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr17_+_25414033 | 0.38 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr13_+_39277178 | 0.38 |
ENSDART00000113259
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr22_-_20105969 | 0.37 |
ENSDART00000088687
|
rxfp3.2b
|
relaxin/insulin-like family peptide receptor 3.2b |
| chr2_+_22659787 | 0.36 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr12_+_24342303 | 0.36 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr17_-_7440397 | 0.36 |
ENSDART00000162597
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr10_-_41450367 | 0.35 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr4_+_5180650 | 0.35 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr15_-_20731297 | 0.35 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr12_-_15620090 | 0.34 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr11_+_45299447 | 0.34 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr8_-_26961779 | 0.34 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr16_-_31622777 | 0.33 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr5_+_4366431 | 0.33 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr19_+_6938289 | 0.32 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr24_-_22702017 | 0.32 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr2_-_44720551 | 0.31 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr13_+_41917606 | 0.31 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr5_+_1965296 | 0.31 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr7_+_31319876 | 0.31 |
ENSDART00000187611
|
fam189a1
|
family with sequence similarity 189, member A1 |
| chr19_-_3303995 | 0.31 |
ENSDART00000105150
|
si:ch211-133n4.9
|
si:ch211-133n4.9 |
| chr20_+_50115335 | 0.30 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr5_+_63785339 | 0.30 |
ENSDART00000050871
|
rgs3b
|
regulator of G protein signaling 3b |
| chr7_+_20031202 | 0.30 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr8_+_25247245 | 0.30 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr14_+_14836468 | 0.30 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr16_+_50741154 | 0.30 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr22_-_15569736 | 0.29 |
ENSDART00000132334
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr21_-_20765338 | 0.29 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr18_-_9046805 | 0.29 |
ENSDART00000134224
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr7_+_21752168 | 0.29 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr20_-_27733683 | 0.29 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr8_-_47329755 | 0.29 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr9_+_42066030 | 0.29 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr19_+_33732188 | 0.28 |
ENSDART00000151192
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_+_20030888 | 0.28 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr6_+_39184236 | 0.28 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr2_+_40294313 | 0.27 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr12_-_6159545 | 0.27 |
ENSDART00000152487
|
rltgr
|
RAMP-like triterpene glycoside receptor |
| chr16_-_29458806 | 0.27 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr14_-_32016615 | 0.27 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr9_+_36314867 | 0.27 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr7_+_29167744 | 0.27 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr9_-_44295071 | 0.27 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr20_+_19212962 | 0.26 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr22_-_17606575 | 0.26 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr6_-_16456093 | 0.26 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr20_-_5369105 | 0.26 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr1_-_22834824 | 0.26 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr5_-_18897482 | 0.25 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr18_-_5875433 | 0.25 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr21_-_2322102 | 0.24 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr9_+_45789887 | 0.24 |
ENSDART00000135202
|
si:dkey-34f9.3
|
si:dkey-34f9.3 |
| chr13_-_18637244 | 0.24 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr24_+_2519761 | 0.24 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr9_-_35557397 | 0.24 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr25_-_21092222 | 0.24 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr2_+_6181383 | 0.23 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr10_+_21559605 | 0.23 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr24_-_8409641 | 0.23 |
ENSDART00000149662
ENSDART00000149025 |
slc35b3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr10_+_23022263 | 0.23 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr11_-_3959477 | 0.23 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr6_+_22597362 | 0.23 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr22_-_38034852 | 0.23 |
ENSDART00000104613
|
pex6
|
peroxisomal biogenesis factor 6 |
| chr23_+_40452157 | 0.23 |
ENSDART00000113106
ENSDART00000140136 |
soga3b
|
SOGA family member 3b |
| chr25_-_8625601 | 0.23 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr13_-_40316367 | 0.23 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr4_-_211714 | 0.22 |
ENSDART00000172566
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr6_+_42819337 | 0.22 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr15_-_14469704 | 0.22 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr8_+_20438884 | 0.22 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr18_+_5875268 | 0.22 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr6_-_44711942 | 0.22 |
ENSDART00000055035
|
cntn3b
|
contactin 3b |
| chr11_-_37411492 | 0.22 |
ENSDART00000166468
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr25_+_17689565 | 0.21 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr8_-_19649617 | 0.21 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr15_+_43093044 | 0.21 |
ENSDART00000141125
|
kcne4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr8_-_30979494 | 0.21 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr1_-_23588996 | 0.21 |
ENSDART00000102600
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr3_+_53773256 | 0.21 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr10_-_6775271 | 0.20 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr18_-_226800 | 0.20 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr13_+_18311410 | 0.20 |
ENSDART00000036718
ENSDART00000132073 |
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr24_-_3783258 | 0.20 |
ENSDART00000139596
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr12_-_3453589 | 0.20 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr19_+_33732487 | 0.20 |
ENSDART00000010294
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_+_42469058 | 0.20 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr5_-_13167097 | 0.20 |
ENSDART00000149700
ENSDART00000030213 |
mapk1
|
mitogen-activated protein kinase 1 |
| chr7_-_35126374 | 0.20 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr21_-_36948 | 0.19 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr12_-_999762 | 0.19 |
ENSDART00000127003
ENSDART00000084076 ENSDART00000152425 |
mettl9
|
methyltransferase like 9 |
| chr16_+_10841163 | 0.19 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr18_+_6126506 | 0.19 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr22_+_3238474 | 0.19 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr5_+_27583445 | 0.19 |
ENSDART00000136488
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr12_+_33038757 | 0.19 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr16_+_36906693 | 0.19 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr14_-_46675850 | 0.19 |
ENSDART00000113285
|
tapt1a
|
transmembrane anterior posterior transformation 1a |
| chr5_-_45894802 | 0.18 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr3_+_17744339 | 0.18 |
ENSDART00000132622
|
znf385c
|
zinc finger protein 385C |
| chr18_+_10784730 | 0.18 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr9_-_403767 | 0.18 |
ENSDART00000167743
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr5_+_44228526 | 0.18 |
ENSDART00000143789
|
TMEM8B
|
si:dkey-84j12.1 |
| chr3_-_40933415 | 0.18 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr16_-_21489514 | 0.17 |
ENSDART00000149525
ENSDART00000148517 ENSDART00000146914 ENSDART00000186493 ENSDART00000193081 ENSDART00000186017 |
mpp6a
|
membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) |
| chr11_-_16975190 | 0.17 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr17_+_19499157 | 0.17 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr11_-_37880492 | 0.17 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr19_-_791016 | 0.17 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr5_-_3839285 | 0.17 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr9_+_32358514 | 0.17 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr22_+_17261801 | 0.17 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr1_+_17376922 | 0.17 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr19_-_5103313 | 0.17 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr19_+_9111550 | 0.17 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr12_+_34827808 | 0.16 |
ENSDART00000105533
|
tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr20_-_22798794 | 0.16 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr21_-_41588129 | 0.16 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr14_-_42231293 | 0.16 |
ENSDART00000185486
|
BX890543.1
|
|
| chr20_-_9199721 | 0.16 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr23_+_21459263 | 0.16 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr7_+_32021669 | 0.16 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr21_+_1378250 | 0.16 |
ENSDART00000186912
|
TCF4
|
transcription factor 4 |
| chr7_+_48999723 | 0.16 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr19_-_30565122 | 0.16 |
ENSDART00000185650
|
hpcal4
|
hippocalcin like 4 |
| chr2_+_205763 | 0.16 |
ENSDART00000160164
ENSDART00000101071 |
zgc:113293
|
zgc:113293 |
| chr3_+_40841897 | 0.16 |
ENSDART00000150081
ENSDART00000126096 |
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr9_+_32872690 | 0.16 |
ENSDART00000020798
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr6_-_57476465 | 0.15 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr3_-_11972516 | 0.15 |
ENSDART00000140123
|
hmox2b
|
heme oxygenase 2b |
| chr5_+_27583117 | 0.15 |
ENSDART00000180340
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr4_+_3455665 | 0.15 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr22_-_38035084 | 0.15 |
ENSDART00000145029
|
pex6
|
peroxisomal biogenesis factor 6 |
| chr15_-_11956981 | 0.15 |
ENSDART00000164163
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr12_+_32729470 | 0.15 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr7_+_29163762 | 0.15 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr21_-_22122312 | 0.15 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr18_+_38191346 | 0.15 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr4_-_5302866 | 0.14 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hey2_hey1+heyl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 1.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.5 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.3 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.6 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.4 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.3 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.5 | GO:0030194 | acute-phase response(GO:0006953) positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.2 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.2 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.7 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 1.0 | GO:0009749 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 0.7 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 1.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.0 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.3 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.2 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 1.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.3 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.0 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.0 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.0 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 0.7 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 1.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.2 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0099583 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0052834 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.0 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |