Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
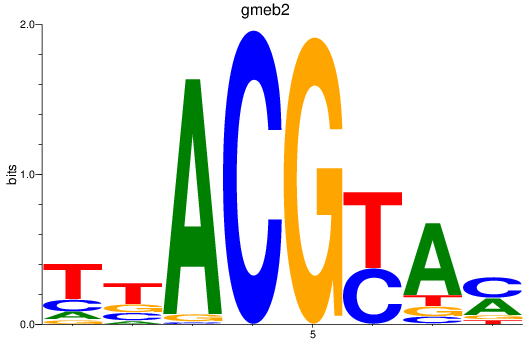
Results for gmeb2
Z-value: 0.73
Transcription factors associated with gmeb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gmeb2
|
ENSDARG00000093240 | si_ch73-302a13.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | dr11_v1_chr23_-_41824460_41824460 | 0.72 | 1.7e-01 | Click! |
Activity profile of gmeb2 motif
Sorted Z-values of gmeb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_27381691 | 0.62 |
ENSDART00000010293
|
serpina10a
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10a |
| chr1_-_38815361 | 0.52 |
ENSDART00000148790
ENSDART00000148572 ENSDART00000149080 |
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr1_+_25801648 | 0.29 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr8_+_1239 | 0.25 |
ENSDART00000067666
ENSDART00000192564 ENSDART00000192612 ENSDART00000190683 |
tmed7
|
transmembrane p24 trafficking protein 7 |
| chr19_-_31402429 | 0.25 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr2_-_32486080 | 0.24 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr16_+_28596555 | 0.24 |
ENSDART00000046209
ENSDART00000141708 |
acbd7
|
acyl-CoA binding domain containing 7 |
| chr11_+_37275448 | 0.23 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr5_+_3501859 | 0.22 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr19_+_46259619 | 0.21 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr11_-_36475124 | 0.21 |
ENSDART00000165203
|
usp48
|
ubiquitin specific peptidase 48 |
| chr8_+_1715 | 0.21 |
ENSDART00000181131
|
tmed7
|
transmembrane p24 trafficking protein 7 |
| chr23_-_35790235 | 0.21 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr22_-_11493236 | 0.20 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr6_-_40744720 | 0.19 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr2_-_21170517 | 0.18 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr3_-_29910547 | 0.18 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr14_-_43000836 | 0.17 |
ENSDART00000162714
|
pcdh10b
|
protocadherin 10b |
| chr8_+_53311965 | 0.17 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr2_-_54387550 | 0.17 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr21_-_43040780 | 0.17 |
ENSDART00000187037
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr22_-_621609 | 0.16 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr12_-_7824291 | 0.16 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr16_+_46111849 | 0.16 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr25_-_36044583 | 0.16 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr24_+_5208171 | 0.16 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr3_-_25814097 | 0.16 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr18_-_40481028 | 0.16 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr4_+_77053467 | 0.15 |
ENSDART00000187643
ENSDART00000150848 |
znf1009
|
zinc finger protein 1009 |
| chr5_+_24245682 | 0.15 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr10_-_14545658 | 0.15 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr9_-_28103097 | 0.15 |
ENSDART00000146284
|
kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr25_-_29988352 | 0.14 |
ENSDART00000067059
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr7_+_69528850 | 0.14 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr19_-_44054930 | 0.14 |
ENSDART00000151084
ENSDART00000150991 ENSDART00000005191 |
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr20_+_8028613 | 0.14 |
ENSDART00000085668
|
prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr25_-_8513255 | 0.14 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr4_-_11810799 | 0.13 |
ENSDART00000014153
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr24_+_20968308 | 0.13 |
ENSDART00000130568
|
drd3
|
dopamine receptor D3 |
| chr2_+_55914699 | 0.13 |
ENSDART00000157905
|
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr15_-_11683529 | 0.13 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr5_-_13076779 | 0.13 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr11_+_43419809 | 0.13 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr2_-_31614277 | 0.12 |
ENSDART00000137054
ENSDART00000131507 ENSDART00000137674 ENSDART00000147041 |
si:ch211-106h4.5
|
si:ch211-106h4.5 |
| chr8_-_25120231 | 0.12 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr1_-_8101495 | 0.12 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr1_+_23274710 | 0.12 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr3_+_32789605 | 0.12 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr4_-_11810365 | 0.12 |
ENSDART00000150529
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr8_-_19467011 | 0.12 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr23_-_38330746 | 0.11 |
ENSDART00000192262
|
FO904868.1
|
|
| chr8_+_31941650 | 0.11 |
ENSDART00000138217
|
htr1aa
|
5-hydroxytryptamine (serotonin) receptor 1A a |
| chr20_+_34717403 | 0.11 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr1_+_49415281 | 0.11 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr20_+_38837238 | 0.11 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr6_-_11780070 | 0.11 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr2_-_3403020 | 0.11 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr18_-_46208581 | 0.11 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr1_-_56223913 | 0.11 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr13_+_3938526 | 0.10 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr3_+_32933663 | 0.10 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr16_+_41015163 | 0.10 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr11_-_41132296 | 0.10 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr17_+_27176243 | 0.10 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr11_+_2506516 | 0.10 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr23_+_2703044 | 0.10 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr12_-_48960308 | 0.10 |
ENSDART00000176247
|
CABZ01112647.1
|
|
| chr7_+_13382852 | 0.10 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr11_+_29770966 | 0.10 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr13_+_2442841 | 0.09 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr14_+_8947282 | 0.09 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr18_+_31410652 | 0.09 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr15_+_5116179 | 0.09 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr13_+_4405282 | 0.09 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr23_+_17899226 | 0.09 |
ENSDART00000079487
|
CU234171.1
|
|
| chr8_+_20455134 | 0.09 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr20_+_25486021 | 0.09 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr2_+_38055529 | 0.09 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr17_-_25831569 | 0.09 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr9_+_19529951 | 0.09 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr8_+_47897734 | 0.09 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr14_-_2358774 | 0.09 |
ENSDART00000164809
|
si:ch73-233f7.5
|
si:ch73-233f7.5 |
| chr3_-_5664123 | 0.09 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr8_-_4618653 | 0.08 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr6_-_51541488 | 0.08 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr13_-_21688176 | 0.08 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr14_-_30808174 | 0.08 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr7_-_39751540 | 0.08 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr1_-_6494384 | 0.08 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr18_+_5917625 | 0.08 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr11_-_18799827 | 0.08 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr2_-_10192459 | 0.08 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr25_+_32473277 | 0.08 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr10_-_15910974 | 0.08 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr4_+_9011825 | 0.08 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr3_+_46764278 | 0.08 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr17_-_6076084 | 0.08 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr24_-_7826489 | 0.08 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr19_+_40026041 | 0.08 |
ENSDART00000017359
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr3_+_46763745 | 0.08 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr7_+_57341805 | 0.08 |
ENSDART00000115222
|
rxfp3.3a2
|
relaxin/insulin-like family peptide receptor 3.3a2 |
| chr19_+_19241372 | 0.08 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr7_-_38570878 | 0.08 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr13_-_36680531 | 0.08 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr9_+_24920677 | 0.08 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_+_56012016 | 0.07 |
ENSDART00000146160
ENSDART00000188702 |
loxl5b
|
lysyl oxidase-like 5b |
| chr16_-_47427016 | 0.07 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr17_-_6076266 | 0.07 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr12_-_20303438 | 0.07 |
ENSDART00000153057
|
rhbdf1b
|
rhomboid 5 homolog 1b (Drosophila) |
| chr20_-_32405440 | 0.07 |
ENSDART00000062978
ENSDART00000153411 |
afg1lb
|
AFG1 like ATPase b |
| chr20_-_30938184 | 0.07 |
ENSDART00000147359
ENSDART00000062552 |
wtap
|
WT1 associated protein |
| chr20_+_2227860 | 0.07 |
ENSDART00000152321
|
tmem200a
|
transmembrane protein 200A |
| chr6_-_17849786 | 0.07 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr12_+_17603528 | 0.07 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr8_-_30944465 | 0.07 |
ENSDART00000128792
ENSDART00000191717 ENSDART00000049944 |
smarcb1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1a |
| chr20_-_44576949 | 0.07 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr1_-_7893808 | 0.07 |
ENSDART00000110154
|
radil
|
Ras association and DIL domains |
| chr3_+_46764022 | 0.07 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr19_-_10395683 | 0.07 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr25_-_28668776 | 0.06 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr7_-_55539738 | 0.06 |
ENSDART00000168721
ENSDART00000013796 ENSDART00000148514 |
aprt
|
adenine phosphoribosyltransferase |
| chr6_+_9163007 | 0.06 |
ENSDART00000183054
ENSDART00000157552 |
zgc:112023
|
zgc:112023 |
| chr17_-_43399896 | 0.06 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr12_+_28956374 | 0.06 |
ENSDART00000033878
|
znf668
|
zinc finger protein 668 |
| chr10_-_41907213 | 0.06 |
ENSDART00000167004
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr22_-_817479 | 0.06 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr5_-_16139600 | 0.06 |
ENSDART00000051644
|
coq5
|
coenzyme Q5, methyltransferase |
| chr15_-_12319065 | 0.06 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr3_-_30186296 | 0.06 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr16_-_20294236 | 0.06 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr14_+_6605675 | 0.06 |
ENSDART00000143179
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr13_+_36680564 | 0.06 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr25_-_36044294 | 0.06 |
ENSDART00000182207
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr10_+_43088270 | 0.05 |
ENSDART00000191221
|
xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr16_-_47426482 | 0.05 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr17_+_135590 | 0.05 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr11_-_44945636 | 0.05 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr7_+_4162994 | 0.05 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr15_+_11683114 | 0.05 |
ENSDART00000168233
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr20_-_45661049 | 0.05 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr12_+_3571770 | 0.05 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr11_+_30000814 | 0.05 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr20_+_25486206 | 0.05 |
ENSDART00000172076
|
hook1
|
hook microtubule-tethering protein 1 |
| chr15_-_43284021 | 0.05 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr9_+_35860975 | 0.05 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr21_-_33995710 | 0.05 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr14_+_10625112 | 0.05 |
ENSDART00000143377
ENSDART00000136480 |
nup62l
|
nucleoporin 62 like |
| chr20_-_9199721 | 0.05 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr8_+_7975745 | 0.05 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr9_+_12948511 | 0.05 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr11_+_8170948 | 0.04 |
ENSDART00000112127
ENSDART00000186267 |
dnase2b
|
deoxyribonuclease II beta |
| chr25_-_2723682 | 0.04 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr8_-_5267442 | 0.04 |
ENSDART00000155816
|
ppp2r2aa
|
protein phosphatase 2, regulatory subunit B, alpha a |
| chr15_-_47338769 | 0.04 |
ENSDART00000164983
ENSDART00000176289 |
anapc15
|
anaphase promoting complex subunit 15 |
| chr1_+_27024068 | 0.04 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr2_+_52049239 | 0.04 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr5_-_24245218 | 0.04 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr6_-_40455109 | 0.04 |
ENSDART00000103878
|
vhl
|
von Hippel-Lindau tumor suppressor |
| chr12_+_33460794 | 0.04 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr4_+_23126558 | 0.04 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr22_+_26798853 | 0.04 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr24_+_39638555 | 0.04 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr9_-_4598883 | 0.04 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr17_-_12058171 | 0.04 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr17_+_1514711 | 0.04 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr25_-_21066136 | 0.04 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr25_-_32464149 | 0.04 |
ENSDART00000152787
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr2_+_32826235 | 0.04 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr13_+_12299997 | 0.04 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr9_+_23049500 | 0.04 |
ENSDART00000012478
|
mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr9_-_23747264 | 0.04 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr23_+_20640484 | 0.04 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr2_-_32826108 | 0.04 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr6_+_46668717 | 0.04 |
ENSDART00000064853
|
oxtrl
|
oxytocin receptor like |
| chr1_+_9860381 | 0.04 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr12_+_11650146 | 0.04 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr3_-_8765165 | 0.04 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr20_+_25563105 | 0.04 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr6_-_10728057 | 0.04 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr17_-_51818659 | 0.03 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr5_-_23596339 | 0.03 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr5_-_54395488 | 0.03 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr4_+_9011448 | 0.03 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr3_+_53156813 | 0.03 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr17_-_37195163 | 0.03 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr25_+_32473433 | 0.03 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr12_-_26153101 | 0.03 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr8_+_13064750 | 0.03 |
ENSDART00000039878
|
sap30bp
|
SAP30 binding protein |
| chr6_-_25201810 | 0.03 |
ENSDART00000168683
|
lrrc8c
|
leucine rich repeat containing 8 VRAC subunit C |
| chr14_-_30141162 | 0.03 |
ENSDART00000053916
|
mtnr1ab
|
melatonin receptor 1A b |
| chr13_-_12486076 | 0.03 |
ENSDART00000146195
|
lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr12_-_27588299 | 0.03 |
ENSDART00000178023
ENSDART00000066282 |
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr7_+_51795667 | 0.03 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr25_+_36045072 | 0.03 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr10_+_29137482 | 0.03 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr19_-_35155722 | 0.03 |
ENSDART00000151924
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr18_+_11970987 | 0.03 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr10_+_21511495 | 0.03 |
ENSDART00000178395
|
BX957322.3
|
|
| chr11_+_11120532 | 0.02 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr1_+_26105141 | 0.02 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
Network of associatons between targets according to the STRING database.
First level regulatory network of gmeb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0098917 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0006168 | adenine salvage(GO:0006168) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.0 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0060137 | parturition(GO:0007567) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0061010 | gall bladder development(GO:0061010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.0 | 0.1 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.1 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.0 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |