Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
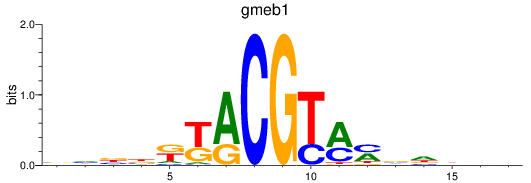
Results for gmeb1
Z-value: 0.44
Transcription factors associated with gmeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gmeb1
|
ENSDARG00000062060 | glucocorticoid modulatory element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gmeb1 | dr11_v1_chr19_+_16064439_16064439 | -0.16 | 8.0e-01 | Click! |
Activity profile of gmeb1 motif
Sorted Z-values of gmeb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_17676745 | 0.54 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr4_-_25485404 | 0.28 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr4_-_25485214 | 0.27 |
ENSDART00000159941
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr16_+_23397785 | 0.20 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr20_+_22666548 | 0.20 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr25_-_34845302 | 0.14 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr16_-_24815091 | 0.11 |
ENSDART00000154269
ENSDART00000131025 |
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr21_-_8153165 | 0.11 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr14_-_8795081 | 0.07 |
ENSDART00000106671
|
tgfb2l
|
transforming growth factor, beta 2, like |
| chr20_+_31287356 | 0.07 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr12_-_48374728 | 0.07 |
ENSDART00000153403
ENSDART00000188117 |
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr5_-_30588539 | 0.06 |
ENSDART00000086686
ENSDART00000159163 ENSDART00000142958 |
c2cd2l
|
c2cd2-like |
| chr14_-_31489410 | 0.05 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr3_-_16000168 | 0.05 |
ENSDART00000160628
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr8_+_10305400 | 0.05 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr3_-_15999501 | 0.05 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr9_+_16241656 | 0.04 |
ENSDART00000154326
|
si:ch211-261p9.4
|
si:ch211-261p9.4 |
| chr12_+_3571770 | 0.04 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr12_+_11650146 | 0.03 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr22_-_621888 | 0.02 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr11_+_35364445 | 0.02 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr10_+_39091716 | 0.02 |
ENSDART00000193072
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr10_+_29138021 | 0.02 |
ENSDART00000025227
ENSDART00000123033 ENSDART00000034242 |
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr21_+_6397463 | 0.01 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr4_+_23126558 | 0.01 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr10_+_26990095 | 0.01 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr4_+_45148652 | 0.01 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr25_-_36044583 | 0.01 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr14_-_21932403 | 0.01 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr25_-_36044294 | 0.00 |
ENSDART00000182207
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr4_-_17642168 | 0.00 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr1_+_19515228 | 0.00 |
ENSDART00000103091
|
clrn2
|
clarin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gmeb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.0 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |