Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
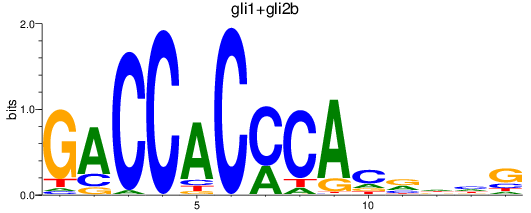
Results for gli1+gli2b
Z-value: 1.26
Transcription factors associated with gli1+gli2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2b
|
ENSDARG00000020884 | GLI family zinc finger 2b |
|
gli1
|
ENSDARG00000101244 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli1 | dr11_v1_chr6_-_59505589_59505589 | -0.37 | 5.5e-01 | Click! |
| gli2b | dr11_v1_chr11_+_43661735_43661735 | -0.28 | 6.5e-01 | Click! |
Activity profile of gli1+gli2b motif
Sorted Z-values of gli1+gli2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_20422661 | 1.27 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr7_+_29952719 | 1.06 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr7_+_29952169 | 1.05 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr12_-_464007 | 1.01 |
ENSDART00000106669
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr18_+_5547185 | 1.00 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr7_+_29951997 | 0.99 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr14_+_15543331 | 0.73 |
ENSDART00000167025
|
si:dkey-203a12.7
|
si:dkey-203a12.7 |
| chr10_+_33171501 | 0.72 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr14_+_15257658 | 0.70 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr14_+_15430991 | 0.61 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr4_-_17741513 | 0.59 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr20_-_54377933 | 0.55 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr14_+_15495088 | 0.54 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr19_+_9004256 | 0.53 |
ENSDART00000186401
ENSDART00000091443 |
si:ch211-81a5.1
|
si:ch211-81a5.1 |
| chr14_+_15231097 | 0.52 |
ENSDART00000172430
|
si:dkey-203a12.3
|
si:dkey-203a12.3 |
| chr14_+_15597049 | 0.51 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr14_+_15331486 | 0.49 |
ENSDART00000172077
ENSDART00000183370 ENSDART00000182467 |
SPINK2
si:dkey-203a12.5
|
si:dkey-203a12.4 si:dkey-203a12.5 |
| chr25_+_30298377 | 0.49 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr1_-_470812 | 0.48 |
ENSDART00000192527
|
zgc:92518
|
zgc:92518 |
| chr12_+_27462225 | 0.46 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr10_+_43188678 | 0.44 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr16_-_13595027 | 0.41 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr20_-_53996193 | 0.41 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr13_-_865193 | 0.36 |
ENSDART00000187053
|
AL929536.7
|
|
| chr8_+_28469054 | 0.34 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr16_-_30885838 | 0.34 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr14_-_29859067 | 0.33 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr20_+_27194833 | 0.32 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr14_-_15171435 | 0.31 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr6_-_3982783 | 0.30 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr22_-_3344613 | 0.29 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr8_-_40327397 | 0.28 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr16_-_43041324 | 0.28 |
ENSDART00000155445
ENSDART00000156836 ENSDART00000154945 |
si:dkey-7j14.6
|
si:dkey-7j14.6 |
| chr21_-_30415524 | 0.27 |
ENSDART00000101036
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr7_+_31871830 | 0.26 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chrM_+_9735 | 0.24 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr14_+_15155684 | 0.22 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr10_-_24391716 | 0.22 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr13_+_228045 | 0.22 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr12_+_35650321 | 0.22 |
ENSDART00000190446
|
BX255898.1
|
|
| chr4_+_10721795 | 0.21 |
ENSDART00000136000
ENSDART00000067253 |
stab2
|
stabilin 2 |
| chr15_-_34056733 | 0.20 |
ENSDART00000170130
ENSDART00000188272 |
VWDE
|
si:dkey-30e9.7 |
| chr7_-_27037990 | 0.20 |
ENSDART00000173561
|
nucb2a
|
nucleobindin 2a |
| chr22_-_22147375 | 0.18 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr10_-_24391030 | 0.17 |
ENSDART00000180013
ENSDART00000177914 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr13_-_17723417 | 0.16 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr7_+_4162994 | 0.16 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr11_+_24314148 | 0.16 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr8_-_14121634 | 0.16 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr10_-_39154594 | 0.16 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr4_-_72101234 | 0.16 |
ENSDART00000167048
|
slco1f1
|
solute carrier organic anion transporter family, member 1F1 |
| chr10_+_15224660 | 0.16 |
ENSDART00000137932
|
sh3bp2
|
SH3-domain binding protein 2 |
| chr12_-_26415499 | 0.15 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr5_+_10046643 | 0.14 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr2_-_44199722 | 0.14 |
ENSDART00000140633
ENSDART00000145728 |
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr24_-_6158933 | 0.13 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr13_-_9598320 | 0.13 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr18_+_25653599 | 0.13 |
ENSDART00000007856
|
fkbp16
|
FK506 binding protein 16 |
| chr11_+_30306606 | 0.13 |
ENSDART00000128276
ENSDART00000190222 |
ugt1b4
|
UDP glucuronosyltransferase 1 family, polypeptide B4 |
| chr21_-_17037907 | 0.13 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr11_+_44300548 | 0.13 |
ENSDART00000191626
|
CABZ01100304.1
|
|
| chr15_-_43287515 | 0.12 |
ENSDART00000155103
|
prss16
|
protease, serine, 16 (thymus) |
| chr3_-_9488350 | 0.12 |
ENSDART00000186707
|
FO904885.2
|
|
| chr16_+_13822137 | 0.11 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr5_+_36439405 | 0.11 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr1_+_59090743 | 0.11 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr6_+_27090800 | 0.10 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr7_+_10701770 | 0.10 |
ENSDART00000167323
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr4_-_77506362 | 0.10 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr15_+_25489406 | 0.10 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr12_+_34273240 | 0.10 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr6_-_30859656 | 0.09 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr15_-_38129845 | 0.09 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr25_+_7461624 | 0.08 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr6_-_436658 | 0.07 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr7_+_10701938 | 0.07 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr22_+_26665422 | 0.07 |
ENSDART00000164994
|
adcy9
|
adenylate cyclase 9 |
| chr1_+_57235896 | 0.07 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr4_-_11077872 | 0.07 |
ENSDART00000150566
|
BX784029.4
|
|
| chr12_-_4756478 | 0.07 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr14_-_45702721 | 0.06 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr23_+_26009266 | 0.06 |
ENSDART00000054025
|
si:dkey-78k11.9
|
si:dkey-78k11.9 |
| chr5_+_40224938 | 0.06 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr7_+_18075504 | 0.06 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr5_+_36513605 | 0.06 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr16_-_12784373 | 0.06 |
ENSDART00000080396
|
foxj2
|
forkhead box J2 |
| chr20_+_51730658 | 0.05 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
| chr4_-_2219705 | 0.05 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr21_-_8370500 | 0.05 |
ENSDART00000055328
|
nek6
|
NIMA-related kinase 6 |
| chr20_+_35484070 | 0.05 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr18_+_14529005 | 0.05 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr4_+_38170708 | 0.05 |
ENSDART00000168900
|
znf1071
|
zinc finger protein 1071 |
| chr17_-_17447899 | 0.05 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr5_-_17876709 | 0.04 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr17_-_722218 | 0.04 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr25_-_34740627 | 0.04 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr13_-_39947335 | 0.04 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr6_+_28796773 | 0.04 |
ENSDART00000168237
ENSDART00000163541 |
tp63
|
tumor protein p63 |
| chr18_-_45617146 | 0.04 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr4_-_5856200 | 0.03 |
ENSDART00000121936
|
slc5a8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr22_+_31207799 | 0.03 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr5_+_52844681 | 0.03 |
ENSDART00000162459
ENSDART00000184914 |
scarb2a
|
scavenger receptor class B, member 2a |
| chr21_-_41873065 | 0.03 |
ENSDART00000014538
|
endou2
|
endonuclease, polyU-specific 2 |
| chr18_-_39636348 | 0.03 |
ENSDART00000129828
|
cyp19a1a
|
cytochrome P450, family 19, subfamily A, polypeptide 1a |
| chr2_+_54086436 | 0.03 |
ENSDART00000174581
|
CU179656.1
|
|
| chr15_+_7992906 | 0.03 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr25_+_32473433 | 0.03 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr17_-_50331351 | 0.03 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr8_+_1065458 | 0.03 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr18_-_40753583 | 0.03 |
ENSDART00000026767
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_+_58877162 | 0.03 |
ENSDART00000122174
|
CABZ01085658.1
|
|
| chr5_-_43959972 | 0.03 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr11_+_14295011 | 0.03 |
ENSDART00000060226
|
si:ch211-262i1.4
|
si:ch211-262i1.4 |
| chr4_+_58016732 | 0.03 |
ENSDART00000165777
|
CT027609.1
|
|
| chr10_+_42691210 | 0.03 |
ENSDART00000193813
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr15_+_32411660 | 0.02 |
ENSDART00000113443
|
si:dkey-285b23.4
|
si:dkey-285b23.4 |
| chr19_-_6988837 | 0.02 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr21_-_21465111 | 0.02 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr3_-_33941319 | 0.02 |
ENSDART00000026090
ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr2_+_30147504 | 0.02 |
ENSDART00000190947
|
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr13_-_1423008 | 0.02 |
ENSDART00000110828
|
znf451
|
zinc finger protein 451 |
| chr10_-_44411032 | 0.02 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr21_-_41873584 | 0.02 |
ENSDART00000188089
|
endou2
|
endonuclease, polyU-specific 2 |
| chr8_-_43847138 | 0.01 |
ENSDART00000148358
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr9_+_17862858 | 0.01 |
ENSDART00000166566
|
dgkh
|
diacylglycerol kinase, eta |
| chr10_+_5645887 | 0.01 |
ENSDART00000171426
|
pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr6_-_11768198 | 0.01 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr4_+_63253425 | 0.01 |
ENSDART00000193510
|
si:ch211-258f1.3
|
si:ch211-258f1.3 |
| chr25_+_245438 | 0.01 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr6_-_10693111 | 0.01 |
ENSDART00000187206
ENSDART00000192319 |
dnah9l
|
dynein, axonemal, heavy polypeptide 9 like |
| chr4_+_5132951 | 0.01 |
ENSDART00000103279
|
ccnd2b
|
cyclin D2, b |
| chr25_+_32474031 | 0.01 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr5_-_22615087 | 0.01 |
ENSDART00000146035
|
zgc:113208
|
zgc:113208 |
| chr19_-_9776075 | 0.01 |
ENSDART00000140332
|
si:dkey-14o18.1
|
si:dkey-14o18.1 |
| chr1_+_59090583 | 0.01 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr4_+_28997595 | 0.01 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr21_+_10739846 | 0.00 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr5_-_25123807 | 0.00 |
ENSDART00000183171
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr1_-_31505144 | 0.00 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr23_+_28582865 | 0.00 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
Network of associatons between targets according to the STRING database.
First level regulatory network of gli1+gli2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.4 | GO:1903428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 0.3 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 1.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 3.0 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.0 | 0.2 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0030237 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 3.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.3 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 3.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.6 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |