Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
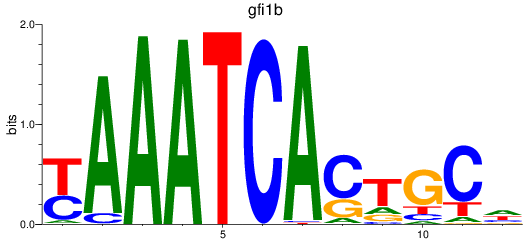
Results for gfi1b
Z-value: 1.52
Transcription factors associated with gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1b
|
ENSDARG00000079947 | growth factor independent 1B transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1b | dr11_v1_chr21_-_17290941_17290941 | -0.50 | 3.9e-01 | Click! |
Activity profile of gfi1b motif
Sorted Z-values of gfi1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_19999623 | 2.28 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr20_+_50852356 | 2.25 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr17_-_12389259 | 2.02 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr23_-_35694461 | 1.84 |
ENSDART00000185884
|
tuba1c
|
tubulin, alpha 1c |
| chr23_-_35694171 | 1.81 |
ENSDART00000077539
|
tuba1c
|
tubulin, alpha 1c |
| chr17_-_12385308 | 1.71 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr17_+_15433518 | 1.62 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 1.60 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr1_+_12763461 | 1.46 |
ENSDART00000159226
ENSDART00000180121 |
pcdh10a
|
protocadherin 10a |
| chr21_+_9628854 | 1.39 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr20_-_29864390 | 1.36 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr3_-_36440705 | 1.35 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr10_-_34871737 | 1.31 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr21_-_4032650 | 1.28 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr6_+_55277419 | 1.18 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr5_-_40734045 | 1.08 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr16_+_43152727 | 1.03 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr19_-_9503473 | 1.01 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr7_-_25895189 | 1.00 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr23_-_29505463 | 0.97 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr7_-_26408472 | 0.96 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr9_+_30108641 | 0.95 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr18_-_16181952 | 0.93 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr4_-_12007404 | 0.93 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr25_-_518656 | 0.93 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr5_-_40510397 | 0.89 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr17_+_9308425 | 0.89 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr2_+_9822319 | 0.89 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr2_+_34967210 | 0.84 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr9_-_3653259 | 0.82 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr6_+_11249706 | 0.80 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr16_+_39159752 | 0.78 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr12_+_21299338 | 0.77 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr24_-_32408404 | 0.77 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr2_+_54482603 | 0.76 |
ENSDART00000130977
ENSDART00000183090 |
mtcl1
|
microtubule crosslinking factor 1 |
| chr10_-_34867401 | 0.76 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr16_-_263658 | 0.75 |
ENSDART00000129303
|
slc6a3
|
solute carrier family 6 (neurotransmitter transporter), member 3 |
| chr5_-_23362602 | 0.73 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr3_-_42086577 | 0.72 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr25_-_3979583 | 0.72 |
ENSDART00000124749
|
myrf
|
myelin regulatory factor |
| chr7_-_51603276 | 0.71 |
ENSDART00000174054
|
nhsl2
|
NHS-like 2 |
| chr23_-_29505645 | 0.71 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr13_+_15816573 | 0.71 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr7_+_22801465 | 0.70 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr2_+_9821757 | 0.69 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr7_-_72208248 | 0.69 |
ENSDART00000108916
|
zmp:0000001168
|
zmp:0000001168 |
| chr6_+_11250033 | 0.68 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr20_-_26042070 | 0.68 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr9_+_34641237 | 0.68 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr6_-_10780698 | 0.66 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr2_+_34967022 | 0.66 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr6_-_12851888 | 0.65 |
ENSDART00000056764
|
bmpr2a
|
bone morphogenetic protein receptor, type II a (serine/threonine kinase) |
| chr7_-_28148310 | 0.63 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr2_-_36925561 | 0.62 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr4_-_20235904 | 0.62 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr16_-_44945224 | 0.62 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr1_-_47089818 | 0.61 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr19_+_29798064 | 0.61 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr5_+_45976130 | 0.60 |
ENSDART00000175670
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr14_-_21218891 | 0.60 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr25_-_3979288 | 0.60 |
ENSDART00000157117
|
myrf
|
myelin regulatory factor |
| chr18_+_27511976 | 0.58 |
ENSDART00000132017
ENSDART00000140781 |
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr5_+_44064764 | 0.58 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr16_+_23282655 | 0.58 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr2_+_31838442 | 0.57 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr2_-_30668580 | 0.57 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr6_+_35362225 | 0.56 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr20_+_28434196 | 0.55 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr14_-_47314340 | 0.55 |
ENSDART00000164851
|
fstl5
|
follistatin-like 5 |
| chr21_+_7605803 | 0.55 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr5_+_43965078 | 0.54 |
ENSDART00000113502
ENSDART00000187143 |
TMEM8B
|
si:dkey-84j12.1 |
| chr22_+_31821815 | 0.54 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr8_-_19395110 | 0.53 |
ENSDART00000014183
|
colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr7_+_28960518 | 0.53 |
ENSDART00000182903
ENSDART00000188250 ENSDART00000191830 |
dok4
|
docking protein 4 |
| chr13_-_42724645 | 0.52 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr4_-_75057322 | 0.52 |
ENSDART00000157935
|
large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr25_-_15049694 | 0.52 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr7_+_25920792 | 0.51 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr12_-_3778848 | 0.51 |
ENSDART00000152128
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr5_+_57773222 | 0.50 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr14_-_47391084 | 0.50 |
ENSDART00000159608
|
fstl5
|
follistatin-like 5 |
| chr23_+_22656477 | 0.50 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr6_+_11250316 | 0.50 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr6_+_46431848 | 0.49 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr11_-_6188413 | 0.49 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr7_+_7019911 | 0.47 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr23_-_10137322 | 0.46 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr13_+_15657911 | 0.46 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr25_-_16818978 | 0.45 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr5_-_36328688 | 0.43 |
ENSDART00000011399
|
efnb1
|
ephrin-B1 |
| chr5_-_62306819 | 0.42 |
ENSDART00000168993
|
spata22
|
spermatogenesis associated 22 |
| chr19_+_21919856 | 0.41 |
ENSDART00000187306
ENSDART00000138544 |
galr1a
|
galanin receptor 1a |
| chr16_-_41667101 | 0.41 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr5_+_60928576 | 0.40 |
ENSDART00000131041
|
doc2b
|
double C2-like domains, beta |
| chr10_-_39153959 | 0.40 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr24_+_26402110 | 0.40 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr19_-_31707892 | 0.40 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr11_-_2297832 | 0.39 |
ENSDART00000158266
|
znf740a
|
zinc finger protein 740a |
| chr21_-_33995710 | 0.39 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr8_-_52715911 | 0.39 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr9_+_44722205 | 0.38 |
ENSDART00000086176
ENSDART00000145271 ENSDART00000132696 |
nckap1
|
NCK-associated protein 1 |
| chr7_+_35191220 | 0.37 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr6_-_12912606 | 0.37 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr17_+_34805897 | 0.36 |
ENSDART00000137090
ENSDART00000077626 |
id2a
|
inhibitor of DNA binding 2a |
| chr20_-_40487208 | 0.36 |
ENSDART00000075070
ENSDART00000142029 |
hsf2
|
heat shock transcription factor 2 |
| chr6_-_32349153 | 0.35 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr3_-_39695856 | 0.35 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr10_+_9159279 | 0.35 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr17_+_24687338 | 0.35 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr7_+_48319916 | 0.35 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr14_-_6943934 | 0.35 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr21_+_25068215 | 0.34 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr8_-_43574935 | 0.32 |
ENSDART00000051139
|
ncor2
|
nuclear receptor corepressor 2 |
| chr3_+_26223376 | 0.31 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr13_+_21779975 | 0.31 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr11_+_25634041 | 0.31 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr17_+_11675362 | 0.31 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr19_-_28130658 | 0.31 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr20_-_43917647 | 0.31 |
ENSDART00000026213
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr16_-_23471387 | 0.30 |
ENSDART00000180899
|
CR533578.1
|
|
| chr6_+_3516926 | 0.30 |
ENSDART00000151607
|
faah
|
fatty acid amide hydrolase |
| chr14_+_49007501 | 0.29 |
ENSDART00000128508
|
zdhhc5b
|
zinc finger, DHHC-type containing 5b |
| chr6_+_3516695 | 0.29 |
ENSDART00000109576
|
faah
|
fatty acid amide hydrolase |
| chr14_+_35023923 | 0.29 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr3_-_12970418 | 0.29 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr13_+_22712406 | 0.28 |
ENSDART00000132847
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr8_+_20495889 | 0.27 |
ENSDART00000138794
ENSDART00000021683 |
csnk1g2b
|
casein kinase 1, gamma 2b |
| chr9_-_459910 | 0.27 |
ENSDART00000162551
|
si:dkey-11f4.16
|
si:dkey-11f4.16 |
| chr4_-_63923523 | 0.26 |
ENSDART00000144330
|
si:dkey-179k24.1
|
si:dkey-179k24.1 |
| chr9_+_19489514 | 0.26 |
ENSDART00000152032
ENSDART00000114256 ENSDART00000190572 ENSDART00000147571 ENSDART00000151918 ENSDART00000152034 |
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr24_+_9412450 | 0.26 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr3_-_39696066 | 0.25 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr11_+_25044082 | 0.25 |
ENSDART00000123263
|
phf20a
|
PHD finger protein 20, a |
| chr13_-_31878043 | 0.25 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr23_-_3721444 | 0.25 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr20_+_13783040 | 0.24 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr22_-_4407871 | 0.23 |
ENSDART00000162523
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr19_+_4912817 | 0.23 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_+_39054478 | 0.23 |
ENSDART00000173825
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr6_+_45494227 | 0.21 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr1_+_47331644 | 0.20 |
ENSDART00000053284
|
bcl9
|
B cell CLL/lymphoma 9 |
| chr2_-_13254821 | 0.20 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr20_+_34596205 | 0.20 |
ENSDART00000138338
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr21_-_131236 | 0.20 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr13_-_31878263 | 0.19 |
ENSDART00000180411
|
syt14a
|
synaptotagmin XIVa |
| chr12_-_26022663 | 0.19 |
ENSDART00000166769
|
bmpr1ab
|
bone morphogenetic protein receptor, type IAb |
| chr5_-_58832332 | 0.19 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr18_-_22735002 | 0.19 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr21_+_39963851 | 0.19 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr21_-_2935455 | 0.19 |
ENSDART00000163963
|
CABZ01023255.1
|
|
| chr4_-_16706776 | 0.18 |
ENSDART00000079461
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr4_-_17642168 | 0.16 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr9_+_6255973 | 0.16 |
ENSDART00000078525
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr14_+_3944826 | 0.16 |
ENSDART00000170167
|
lrp2bp
|
LRP2 binding protein |
| chr7_-_35036770 | 0.16 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr7_-_33684632 | 0.16 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr7_-_35083184 | 0.16 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr4_+_6833583 | 0.15 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr8_-_49495584 | 0.15 |
ENSDART00000141691
|
opn7d
|
opsin 7, group member d |
| chr12_+_38563073 | 0.14 |
ENSDART00000009172
|
ttyh2
|
tweety family member 2 |
| chr12_-_4220713 | 0.13 |
ENSDART00000129427
|
vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr2_-_15040345 | 0.13 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr12_+_4220353 | 0.13 |
ENSDART00000133675
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr19_-_12193622 | 0.13 |
ENSDART00000041960
|
ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr21_-_33995213 | 0.13 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr19_-_31584444 | 0.12 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr4_-_27897160 | 0.12 |
ENSDART00000066924
ENSDART00000066925 ENSDART00000193020 |
tbc1d22a
|
TBC1 domain family, member 22a |
| chr2_-_31800521 | 0.12 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr20_+_6535176 | 0.12 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr6_+_37655078 | 0.11 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr2_-_13254594 | 0.11 |
ENSDART00000155671
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr7_+_13609457 | 0.11 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr14_-_45512807 | 0.10 |
ENSDART00000173172
|
si:ch211-114c17.1
|
si:ch211-114c17.1 |
| chr25_+_17313568 | 0.09 |
ENSDART00000125459
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr21_+_27370671 | 0.09 |
ENSDART00000009234
ENSDART00000142071 |
rbm14a
|
RNA binding motif protein 14a |
| chr2_-_293793 | 0.09 |
ENSDART00000082091
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr3_+_51684963 | 0.08 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr15_-_41290415 | 0.08 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr5_+_51909740 | 0.07 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr2_+_37836821 | 0.07 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr16_+_10329701 | 0.07 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr16_-_54978981 | 0.06 |
ENSDART00000154023
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr8_+_23615132 | 0.06 |
ENSDART00000099769
|
ccdc22
|
coiled-coil domain containing 22 |
| chr5_+_36439405 | 0.06 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr24_-_6628359 | 0.06 |
ENSDART00000169731
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr11_-_42396302 | 0.06 |
ENSDART00000165624
|
slmapa
|
sarcolemma associated protein a |
| chr2_-_293036 | 0.06 |
ENSDART00000171629
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr20_+_27464721 | 0.05 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr4_+_6833735 | 0.05 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr13_-_24396199 | 0.05 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr16_+_26612401 | 0.05 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr17_-_37195163 | 0.04 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr6_+_50393047 | 0.04 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr6_-_3924723 | 0.04 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr23_-_18415872 | 0.04 |
ENSDART00000135430
|
fam120c
|
family with sequence similarity 120C |
| chr21_-_17603182 | 0.04 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr4_-_13567387 | 0.03 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr24_-_23716097 | 0.03 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr3_-_19367081 | 0.03 |
ENSDART00000191369
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr16_+_25608778 | 0.03 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.6 | 1.7 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.3 | 1.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.3 | 3.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 0.8 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.2 | 0.9 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 2.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.9 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.2 | 0.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 1.3 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.1 | 0.4 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.4 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.5 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.4 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 1.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.5 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.3 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.2 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.2 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.2 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 1.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.6 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 1.4 | GO:1903052 | positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) |
| 0.0 | 0.4 | GO:0031280 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.5 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.6 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.2 | GO:2000253 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.8 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.6 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.0 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 1.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 2.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.2 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.3 | 0.8 | GO:0005334 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.2 | 3.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 0.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 0.5 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.6 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 1.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 4.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 3.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |