Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
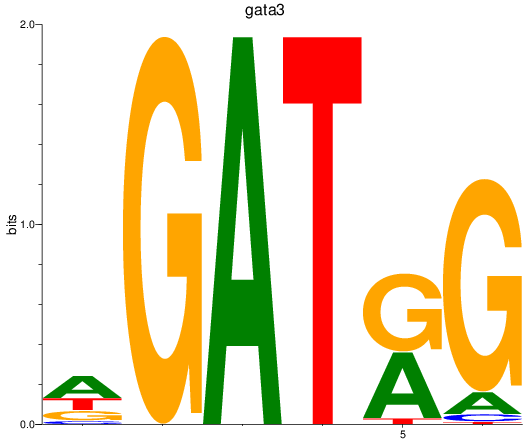
Results for gata3
Z-value: 2.99
Transcription factors associated with gata3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata3
|
ENSDARG00000016526 | GATA binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata3 | dr11_v1_chr4_-_25064510_25064510 | 0.16 | 8.0e-01 | Click! |
Activity profile of gata3 motif
Sorted Z-values of gata3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_49159207 | 5.43 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr7_-_72067475 | 5.01 |
ENSDART00000017763
|
LO018380.1
|
|
| chr7_+_13382852 | 4.99 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr21_+_24287403 | 4.60 |
ENSDART00000111169
|
cadm1a
|
cell adhesion molecule 1a |
| chr22_+_635813 | 4.16 |
ENSDART00000179067
|
CU856139.1
|
|
| chr12_+_5189776 | 3.67 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr21_-_43079161 | 3.47 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr20_+_9828769 | 3.44 |
ENSDART00000160480
ENSDART00000053844 ENSDART00000080691 |
stxbp6l
|
syntaxin binding protein 6 (amisyn), like |
| chr9_-_35557397 | 3.39 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr19_+_12444943 | 3.27 |
ENSDART00000135706
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr16_-_29437373 | 3.09 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr17_-_21280185 | 2.76 |
ENSDART00000123198
|
hspa12a
|
heat shock protein 12A |
| chr19_-_32710922 | 2.55 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr5_-_2282256 | 2.29 |
ENSDART00000064012
|
ca4a
|
carbonic anhydrase IV a |
| chr15_+_40008370 | 2.29 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr14_+_50770537 | 2.15 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr25_+_32157326 | 2.07 |
ENSDART00000112588
|
tjp1b
|
tight junction protein 1b |
| chr9_-_51370293 | 2.04 |
ENSDART00000084806
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr13_-_46991577 | 2.02 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr10_-_2527342 | 1.97 |
ENSDART00000184168
|
CU856539.1
|
|
| chr19_-_36675023 | 1.95 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr7_-_13381129 | 1.94 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr2_+_9552456 | 1.90 |
ENSDART00000056896
|
dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr11_-_1131569 | 1.90 |
ENSDART00000173228
|
slc6a11b
|
solute carrier family 6 (neurotransmitter transporter), member 11b |
| chr25_-_11088839 | 1.86 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr11_+_34824099 | 1.83 |
ENSDART00000037017
ENSDART00000146944 |
slc38a3a
|
solute carrier family 38, member 3a |
| chr6_-_29105727 | 1.81 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr3_-_15352303 | 1.78 |
ENSDART00000104338
ENSDART00000145919 ENSDART00000132135 |
pitpnbl
|
phosphatidylinositol transfer protein, beta, like |
| chr25_+_7461624 | 1.78 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr21_+_43561650 | 1.77 |
ENSDART00000085071
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr16_-_35532937 | 1.77 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr2_+_53204750 | 1.77 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr17_-_37214196 | 1.76 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr20_-_4766645 | 1.70 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr15_-_16704417 | 1.62 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr8_-_22698651 | 1.59 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr10_-_31175744 | 1.52 |
ENSDART00000191728
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr7_-_48173440 | 1.49 |
ENSDART00000124075
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr17_+_16873417 | 1.48 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr7_-_37622370 | 1.42 |
ENSDART00000173523
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr4_+_6643421 | 1.38 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr7_-_24699985 | 1.37 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr15_-_15357178 | 1.36 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr23_-_41762956 | 1.34 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr16_-_52736549 | 1.34 |
ENSDART00000146607
|
azin1a
|
antizyme inhibitor 1a |
| chr25_-_19433244 | 1.34 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr17_+_5768608 | 1.32 |
ENSDART00000157039
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr5_+_44654535 | 1.29 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr13_+_35745572 | 1.28 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr22_+_15507218 | 1.27 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr5_+_44655148 | 1.25 |
ENSDART00000124059
|
dapk1
|
death-associated protein kinase 1 |
| chr12_+_5458462 | 1.22 |
ENSDART00000059735
ENSDART00000153823 |
drd6b
|
dopamine receptor D6b |
| chr10_+_42589391 | 1.22 |
ENSDART00000067689
ENSDART00000075259 |
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr8_+_2438638 | 1.21 |
ENSDART00000141263
|
ENKD1
|
si:ch211-220d9.3 |
| chr17_+_28882977 | 1.18 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr12_+_28986308 | 1.15 |
ENSDART00000153178
|
si:rp71-1c10.8
|
si:rp71-1c10.8 |
| chr15_+_41815703 | 1.14 |
ENSDART00000059508
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr18_+_16963881 | 1.13 |
ENSDART00000147583
|
si:ch211-242e8.1
|
si:ch211-242e8.1 |
| chr4_+_4267451 | 1.12 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr14_+_6546516 | 1.12 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr6_+_2174082 | 1.11 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr9_+_29603649 | 1.05 |
ENSDART00000140477
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr17_+_15033822 | 1.05 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr5_-_11943750 | 1.05 |
ENSDART00000074979
|
rnft2
|
ring finger protein, transmembrane 2 |
| chr15_+_31735931 | 1.03 |
ENSDART00000185681
ENSDART00000149137 |
rxfp2b
|
relaxin/insulin-like family peptide receptor 2b |
| chr24_+_27548474 | 1.01 |
ENSDART00000105774
ENSDART00000123368 |
ek1
|
eph-like kinase 1 |
| chr11_+_34824262 | 1.00 |
ENSDART00000103157
|
slc38a3a
|
solute carrier family 38, member 3a |
| chr4_-_4706893 | 1.00 |
ENSDART00000093005
|
CABZ01020835.1
|
|
| chr23_-_41762797 | 0.99 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr4_-_14315855 | 0.99 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr6_+_37623693 | 0.98 |
ENSDART00000144812
ENSDART00000182709 |
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr17_-_8173380 | 0.98 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr22_+_15438513 | 0.97 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr14_-_32016615 | 0.97 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr22_+_15438872 | 0.96 |
ENSDART00000139800
|
gpc5b
|
glypican 5b |
| chr18_-_34143189 | 0.94 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr16_-_13730152 | 0.92 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr6_-_15604157 | 0.92 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr11_-_36020005 | 0.91 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr6_-_15604417 | 0.90 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr17_+_23255365 | 0.89 |
ENSDART00000180277
|
AL935174.5
|
|
| chr18_-_6460102 | 0.89 |
ENSDART00000137037
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr15_-_33896159 | 0.87 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr21_-_43022048 | 0.84 |
ENSDART00000138329
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr20_-_48061351 | 0.83 |
ENSDART00000164962
|
prep
|
prolyl endopeptidase |
| chr2_+_47623202 | 0.83 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr20_-_35841570 | 0.82 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr17_-_26604549 | 0.82 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr23_-_16692312 | 0.82 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr11_+_1628538 | 0.79 |
ENSDART00000154967
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr14_+_7140997 | 0.79 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr18_-_25051846 | 0.78 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr7_+_24049776 | 0.78 |
ENSDART00000166559
|
efs
|
embryonal Fyn-associated substrate |
| chr9_+_38962017 | 0.78 |
ENSDART00000140436
|
map2
|
microtubule-associated protein 2 |
| chr1_-_23557877 | 0.77 |
ENSDART00000145942
|
fam184b
|
family with sequence similarity 184, member B |
| chr7_+_50053339 | 0.77 |
ENSDART00000174308
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr6_-_9792004 | 0.76 |
ENSDART00000081129
|
cdk15
|
cyclin-dependent kinase 15 |
| chr23_-_28423222 | 0.75 |
ENSDART00000182058
|
c1ql4a
|
complement component 1, q subcomponent-like 4 |
| chr18_-_20822045 | 0.75 |
ENSDART00000100743
|
cttnbp2
|
cortactin binding protein 2 |
| chr11_+_25735478 | 0.74 |
ENSDART00000103566
|
si:dkey-183j2.10
|
si:dkey-183j2.10 |
| chr4_+_4889257 | 0.73 |
ENSDART00000175651
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr7_-_52334840 | 0.73 |
ENSDART00000174173
|
CR938716.1
|
|
| chr10_+_37182626 | 0.71 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr20_-_15922986 | 0.71 |
ENSDART00000189421
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr14_-_30642819 | 0.71 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr15_-_25527580 | 0.70 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr3_-_21137362 | 0.70 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr9_+_52411530 | 0.69 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr18_+_2837563 | 0.68 |
ENSDART00000171495
ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr15_+_30126971 | 0.68 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr19_+_7552699 | 0.67 |
ENSDART00000180788
ENSDART00000115058 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr12_+_38770654 | 0.67 |
ENSDART00000155367
|
kif19
|
kinesin family member 19 |
| chr25_+_33063762 | 0.66 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr17_+_23554932 | 0.66 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr11_-_42554290 | 0.66 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr7_+_71955486 | 0.65 |
ENSDART00000189349
|
CABZ01071171.1
|
Danio rerio low density lipoprotein receptor-related protein 4 (lrp4), mRNA. |
| chr10_+_22003750 | 0.65 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr20_-_47422469 | 0.65 |
ENSDART00000021341
|
kif3ca
|
kinesin family member 3Ca |
| chr4_-_25485404 | 0.65 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr11_-_2594045 | 0.64 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr8_+_26859639 | 0.63 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr25_+_16880990 | 0.63 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr20_-_40119872 | 0.62 |
ENSDART00000191569
|
nkain2
|
sodium/potassium transporting ATPase interacting 2 |
| chr4_-_20051141 | 0.62 |
ENSDART00000066963
|
atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr6_+_54142311 | 0.61 |
ENSDART00000154115
|
hmga1b
|
high mobility group AT-hook 1b |
| chr10_+_37181780 | 0.61 |
ENSDART00000187625
|
ksr1a
|
kinase suppressor of ras 1a |
| chr23_-_19953089 | 0.60 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr18_+_3169579 | 0.60 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr19_-_45985989 | 0.60 |
ENSDART00000165897
|
si:ch211-153f2.3
|
si:ch211-153f2.3 |
| chr2_+_22694382 | 0.59 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr24_-_38079261 | 0.58 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr17_+_26569601 | 0.58 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr14_-_36397768 | 0.58 |
ENSDART00000185199
ENSDART00000052562 |
spata4
|
spermatogenesis associated 4 |
| chr7_-_37622153 | 0.58 |
ENSDART00000052379
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr25_-_24074500 | 0.57 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr21_+_33249478 | 0.57 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr3_-_40768548 | 0.57 |
ENSDART00000004923
|
smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr6_-_43092175 | 0.57 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr22_+_21398508 | 0.56 |
ENSDART00000089408
ENSDART00000186091 |
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr12_+_36413886 | 0.56 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr2_+_5621529 | 0.56 |
ENSDART00000144187
|
fgf12a
|
fibroblast growth factor 12a |
| chr7_-_52334429 | 0.55 |
ENSDART00000187372
|
CR938716.1
|
|
| chr12_+_33151246 | 0.55 |
ENSDART00000162681
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr25_-_29987839 | 0.55 |
ENSDART00000154088
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr10_-_36592486 | 0.55 |
ENSDART00000131693
|
slc1a3b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3b |
| chr20_-_47550577 | 0.54 |
ENSDART00000187361
|
CABZ01053323.1
|
|
| chr10_+_45128375 | 0.54 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr8_-_21268303 | 0.53 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr3_-_1317290 | 0.53 |
ENSDART00000047094
|
LO018552.1
|
|
| chr14_+_44545092 | 0.52 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr4_-_25485214 | 0.52 |
ENSDART00000159941
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr23_+_34321237 | 0.52 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr2_+_48303142 | 0.52 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr19_+_30662529 | 0.52 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr5_+_58397646 | 0.52 |
ENSDART00000180759
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr21_+_9689103 | 0.51 |
ENSDART00000165132
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr23_-_6660985 | 0.51 |
ENSDART00000162405
|
CR450824.2
|
|
| chr20_-_40717900 | 0.51 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr3_-_33901483 | 0.50 |
ENSDART00000144774
ENSDART00000138765 |
cacna1aa
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a |
| chr24_-_24281792 | 0.49 |
ENSDART00000146482
ENSDART00000018420 |
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr21_+_9576176 | 0.49 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr20_-_44557037 | 0.49 |
ENSDART00000140995
|
mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr17_-_17447899 | 0.48 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr24_+_2519761 | 0.48 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr3_-_46818001 | 0.47 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr16_+_14029283 | 0.47 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr7_+_72882446 | 0.47 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr10_+_44581378 | 0.47 |
ENSDART00000190331
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr6_+_51773873 | 0.47 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr2_+_50862527 | 0.46 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr1_+_1941031 | 0.46 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr4_+_17336557 | 0.46 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr3_-_19200571 | 0.46 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr3_-_5664123 | 0.45 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr18_-_14734678 | 0.45 |
ENSDART00000142462
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr5_+_11943792 | 0.45 |
ENSDART00000114873
|
zgc:110063
|
zgc:110063 |
| chr8_+_17078692 | 0.45 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr2_+_6181383 | 0.45 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr9_-_8670158 | 0.45 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr21_+_25643880 | 0.44 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr7_+_44484853 | 0.44 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr5_-_43867045 | 0.44 |
ENSDART00000147252
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr2_+_30144979 | 0.44 |
ENSDART00000129365
ENSDART00000130344 |
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr8_+_3820134 | 0.44 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr2_-_37059966 | 0.44 |
ENSDART00000137967
|
diras1b
|
DIRAS family, GTP-binding RAS-like 1b |
| chr8_+_11993150 | 0.44 |
ENSDART00000132492
ENSDART00000191519 ENSDART00000114432 |
ntng2a
|
netrin g2a |
| chr3_+_17547532 | 0.43 |
ENSDART00000175485
ENSDART00000153763 |
kcnh4a
|
potassium voltage-gated channel, subfamily H (eag-related), member 4a |
| chr14_-_7888748 | 0.43 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr18_+_28102620 | 0.43 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr17_-_16965809 | 0.43 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr20_-_34801181 | 0.43 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr3_+_23247325 | 0.43 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr19_-_874888 | 0.43 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr18_+_25003207 | 0.42 |
ENSDART00000099476
ENSDART00000132285 |
fam174b
|
family with sequence similarity 174, member B |
| chr3_-_21242460 | 0.42 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr11_+_39672874 | 0.42 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr2_-_6182098 | 0.42 |
ENSDART00000156167
|
si:ch73-182a11.2
|
si:ch73-182a11.2 |
| chr18_-_15101193 | 0.42 |
ENSDART00000091742
|
btbd11b
|
BTB (POZ) domain containing 11b |
| chr4_+_21129752 | 0.42 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr7_+_529522 | 0.42 |
ENSDART00000190811
|
nrxn2b
|
neurexin 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of gata3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.7 | 2.9 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.7 | 2.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.6 | 2.3 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.5 | 2.0 | GO:0048242 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.5 | 2.0 | GO:1900181 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.5 | 3.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.4 | 1.8 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 1.4 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.3 | 0.8 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.2 | 1.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.2 | 1.3 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 1.2 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.2 | 0.6 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.2 | 0.5 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 0.7 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.2 | 0.9 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.2 | 1.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.2 | 0.9 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.4 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.4 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 1.1 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.8 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.1 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.1 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.1 | 2.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.2 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 1.6 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.3 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 0.5 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 3.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.6 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.6 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.7 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.4 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.3 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.3 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.4 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.1 | 1.0 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 2.2 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 1.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 3.6 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 0.4 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 1.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 4.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 2.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.1 | GO:0033337 | dorsal fin development(GO:0033337) |
| 0.1 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.1 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.7 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.2 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.8 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 1.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 1.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 3.4 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.2 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 1.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.6 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.3 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.2 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0046951 | cellular ketone body metabolic process(GO:0046950) ketone body biosynthetic process(GO:0046951) ketone body metabolic process(GO:1902224) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.3 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.2 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 0.4 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 3.1 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 1.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 2.3 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 1.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 3.9 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.1 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.0 | 0.3 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 1.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 2.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.8 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.9 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.7 | GO:0006885 | regulation of pH(GO:0006885) regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.3 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 1.3 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.7 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 1.5 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.1 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0061182 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.4 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.2 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.1 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.4 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:0033353 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) S-adenosylmethionine cycle(GO:0033353) S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 1.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 1.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 3.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.0 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.2 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.0 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.4 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.2 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.4 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.0 | GO:1900180 | regulation of protein localization to nucleus(GO:1900180) |
| 0.0 | 2.8 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0044107 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.3 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.0 | 0.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.4 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.2 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:2000223 | specification of organ position(GO:0010159) regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.1 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 1.0 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 3.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 3.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 3.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 3.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.1 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 13.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 5.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.4 | GO:0097014 | ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 2.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 11.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.7 | 3.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.5 | 2.9 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.5 | 5.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 1.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 1.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 2.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 3.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 1.0 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 2.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.3 | GO:0070699 | adrenergic receptor binding(GO:0031690) beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.2 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.4 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 4.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 1.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 1.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 3.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 2.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 4.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 5.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.2 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.0 | GO:0004904 | interferon receptor activity(GO:0004904) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 5.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 2.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.7 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |