Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
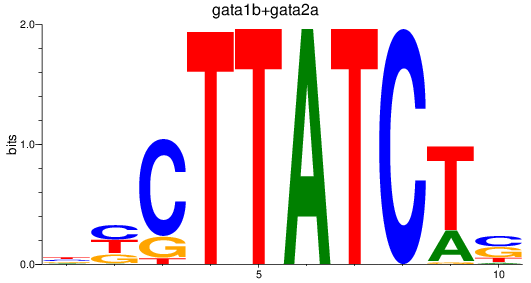
Results for gata1b+gata2a
Z-value: 2.60
Transcription factors associated with gata1b+gata2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata1b
|
ENSDARG00000059130 | GATA binding protein 1b |
|
gata2a
|
ENSDARG00000059327 | GATA binding protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata1b | dr11_v1_chr8_-_7474997_7474997 | -0.76 | 1.3e-01 | Click! |
| gata2a | dr11_v1_chr11_-_3860199_3860199 | -0.27 | 6.7e-01 | Click! |
Activity profile of gata1b+gata2a motif
Sorted Z-values of gata1b+gata2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_7379728 | 4.71 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr16_+_12611362 | 3.81 |
ENSDART00000055160
|
il11a
|
interleukin 11a |
| chr25_-_28674739 | 3.78 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr2_-_24289641 | 3.35 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr5_-_64168415 | 3.14 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr21_-_40880317 | 2.68 |
ENSDART00000100054
ENSDART00000137696 |
elnb
|
elastin b |
| chr2_+_4207209 | 2.57 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr8_-_18537866 | 2.57 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr20_+_40150612 | 2.04 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr5_+_21211135 | 2.03 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr3_+_55105066 | 2.02 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr7_-_73752955 | 1.97 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr2_-_51794472 | 1.94 |
ENSDART00000186652
|
BX908782.3
|
|
| chr8_-_40464935 | 1.88 |
ENSDART00000040013
|
myl7
|
myosin, light chain 7, regulatory |
| chr7_-_41858513 | 1.88 |
ENSDART00000109918
|
mylk3
|
myosin light chain kinase 3 |
| chr23_-_5685023 | 1.85 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr13_+_22480496 | 1.85 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr7_-_41851605 | 1.72 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr13_+_22479988 | 1.68 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr18_+_18000695 | 1.52 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr8_-_50287949 | 1.51 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr20_+_23625387 | 1.48 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr20_+_33981946 | 1.48 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr5_+_23151169 | 1.48 |
ENSDART00000125638
|
tbx5b
|
T-box 5b |
| chr16_+_26449615 | 1.43 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr19_+_37509638 | 1.42 |
ENSDART00000139999
|
si:ch211-250g4.3
|
si:ch211-250g4.3 |
| chr24_-_39858710 | 1.38 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr19_+_46158078 | 1.29 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr19_+_8606883 | 1.28 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr24_+_26276805 | 1.23 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr3_+_55100045 | 1.23 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr7_+_12835048 | 1.23 |
ENSDART00000016465
|
cx36.7
|
connexin 36.7 |
| chr18_-_16795262 | 1.19 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr10_-_17484762 | 1.18 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr13_+_10945337 | 1.00 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr24_-_2843107 | 0.99 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr2_-_38261272 | 0.98 |
ENSDART00000143743
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr20_-_25533739 | 0.98 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr16_-_24945337 | 0.96 |
ENSDART00000154004
|
has1
|
hyaluronan synthase 1 |
| chr22_+_5120033 | 0.95 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr15_-_43164591 | 0.93 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr7_+_44445595 | 0.93 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr20_-_26066020 | 0.92 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr21_+_19547806 | 0.91 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr11_-_1400507 | 0.90 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr11_-_25418856 | 0.89 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr8_+_31872992 | 0.89 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr20_-_33705044 | 0.87 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr4_+_1530287 | 0.87 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr24_-_12958668 | 0.86 |
ENSDART00000178982
|
FITM1 (1 of many)
|
Danio rerio fat storage inducing transmembrane protein 1 (LOC792443), mRNA. |
| chr20_+_33875256 | 0.85 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr17_-_14671098 | 0.84 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr22_+_5118361 | 0.82 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr16_-_17300030 | 0.82 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr18_-_6803424 | 0.81 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr21_-_11834452 | 0.80 |
ENSDART00000081652
|
rfesd
|
Rieske (Fe-S) domain containing |
| chr16_-_24815091 | 0.79 |
ENSDART00000154269
ENSDART00000131025 |
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr18_-_42830563 | 0.79 |
ENSDART00000191488
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr21_-_25756119 | 0.78 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr13_-_10945288 | 0.77 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr7_+_35075847 | 0.77 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr12_+_23850661 | 0.76 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr19_+_20778011 | 0.76 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr11_+_31285127 | 0.74 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr23_+_25856541 | 0.74 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr5_+_15203421 | 0.73 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr1_+_36674584 | 0.72 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr1_-_9109699 | 0.70 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr18_+_48428126 | 0.70 |
ENSDART00000137978
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr22_-_36856405 | 0.68 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr8_+_37700090 | 0.68 |
ENSDART00000187885
ENSDART00000127633 |
adrb3a
|
adrenoceptor beta 3a |
| chr7_-_15252540 | 0.68 |
ENSDART00000173072
ENSDART00000125258 |
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr10_+_26612321 | 0.68 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr15_+_26600611 | 0.66 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr1_-_38816685 | 0.66 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr6_-_609880 | 0.65 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr20_-_33704753 | 0.64 |
ENSDART00000157427
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr7_+_9904627 | 0.63 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr5_-_51166025 | 0.61 |
ENSDART00000097471
|
card9
|
caspase recruitment domain family, member 9 |
| chr12_-_32421046 | 0.60 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr6_-_49078263 | 0.60 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr7_+_17120811 | 0.60 |
ENSDART00000147140
|
prmt3
|
protein arginine methyltransferase 3 |
| chr19_-_46777963 | 0.59 |
ENSDART00000046857
|
abrab
|
actin binding Rho activating protein b |
| chr21_+_12010505 | 0.58 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr3_-_34547000 | 0.58 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr14_+_26546175 | 0.57 |
ENSDART00000105932
|
si:dkeyp-110e4.11
|
si:dkeyp-110e4.11 |
| chr24_+_19518570 | 0.56 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr3_+_16762483 | 0.56 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr20_+_25879826 | 0.55 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr24_+_19518303 | 0.55 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr13_+_835390 | 0.55 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr17_+_39242437 | 0.54 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr15_-_21877726 | 0.53 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr8_+_26059677 | 0.53 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr13_-_25774183 | 0.53 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr10_+_10801719 | 0.52 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr5_-_42272517 | 0.52 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr13_+_24851102 | 0.50 |
ENSDART00000187637
|
si:dkey-24f15.2
|
si:dkey-24f15.2 |
| chr9_+_343062 | 0.50 |
ENSDART00000164012
ENSDART00000164365 |
bpifcl
|
BPI fold containing family C, like |
| chr17_+_34206167 | 0.49 |
ENSDART00000136167
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr10_+_10801564 | 0.49 |
ENSDART00000027026
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr18_+_48428713 | 0.49 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr8_+_6576940 | 0.48 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr18_+_33550667 | 0.48 |
ENSDART00000141246
ENSDART00000136070 |
si:dkey-47k20.1
|
si:dkey-47k20.1 |
| chr11_-_28614608 | 0.46 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr23_+_36115541 | 0.45 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr13_+_844150 | 0.45 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr23_-_35483163 | 0.45 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr5_-_8817841 | 0.44 |
ENSDART00000170921
|
fgf10b
|
fibroblast growth factor 10b |
| chr14_+_30568961 | 0.44 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr17_+_8175998 | 0.44 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr2_+_39021282 | 0.43 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr6_-_30485009 | 0.43 |
ENSDART00000025698
|
zgc:153311
|
zgc:153311 |
| chr9_-_42871756 | 0.42 |
ENSDART00000191396
|
ttn.1
|
titin, tandem duplicate 1 |
| chr25_-_35102781 | 0.42 |
ENSDART00000180881
ENSDART00000153747 |
si:dkey-108k21.24
|
si:dkey-108k21.24 |
| chr6_-_40697585 | 0.42 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr13_-_37103126 | 0.40 |
ENSDART00000135935
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr8_+_23485079 | 0.40 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr12_-_29301022 | 0.40 |
ENSDART00000187826
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr25_+_18564266 | 0.39 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr5_+_26248380 | 0.39 |
ENSDART00000079049
|
si:ch211-214j8.1
|
si:ch211-214j8.1 |
| chr14_+_32839535 | 0.38 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr14_+_2487672 | 0.38 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr14_+_29780113 | 0.38 |
ENSDART00000173195
|
zgc:153146
|
zgc:153146 |
| chr3_+_50602866 | 0.38 |
ENSDART00000190069
ENSDART00000153921 |
gsg1l2a
|
gsg1-like 2a |
| chr13_+_24679674 | 0.38 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr23_+_25708787 | 0.37 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr16_-_24668620 | 0.37 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr24_-_25691020 | 0.37 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr12_-_20350629 | 0.37 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr8_-_40327397 | 0.37 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr24_+_2843268 | 0.36 |
ENSDART00000170529
|
si:ch211-152c8.2
|
si:ch211-152c8.2 |
| chr5_-_68795063 | 0.36 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr4_-_20511595 | 0.36 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr11_-_34577034 | 0.35 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr6_-_39275793 | 0.34 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr14_+_23184517 | 0.34 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr3_-_26204867 | 0.33 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr15_+_29709382 | 0.33 |
ENSDART00000099956
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr13_-_40282770 | 0.32 |
ENSDART00000140875
|
zgc:123010
|
zgc:123010 |
| chr12_+_6391243 | 0.32 |
ENSDART00000152765
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr13_-_23031803 | 0.32 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr1_+_26667872 | 0.32 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr24_+_14801844 | 0.31 |
ENSDART00000141620
|
pi15a
|
peptidase inhibitor 15a |
| chr20_-_35554451 | 0.31 |
ENSDART00000189085
|
adgrf7
|
adhesion G protein-coupled receptor F7 |
| chr16_-_5612480 | 0.31 |
ENSDART00000157944
|
CR848788.1
|
|
| chr20_+_1329509 | 0.31 |
ENSDART00000017791
ENSDART00000136669 |
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr22_+_30112390 | 0.31 |
ENSDART00000191715
|
add3a
|
adducin 3 (gamma) a |
| chr2_+_38261748 | 0.30 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr5_-_37103487 | 0.30 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr9_-_45149695 | 0.30 |
ENSDART00000162033
|
CU467837.1
|
|
| chr5_-_26247973 | 0.29 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr19_+_43523690 | 0.29 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr3_-_33395233 | 0.29 |
ENSDART00000167349
|
si:dkey-283b1.6
|
si:dkey-283b1.6 |
| chr13_+_24263049 | 0.29 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr7_-_51102479 | 0.28 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr1_+_9954489 | 0.28 |
ENSDART00000005895
ENSDART00000183003 |
pdia2
|
protein disulfide isomerase family A, member 2 |
| chr3_+_28502419 | 0.28 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr6_+_58915889 | 0.28 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr14_+_21686207 | 0.28 |
ENSDART00000034438
|
ran
|
RAN, member RAS oncogene family |
| chr7_-_68419830 | 0.27 |
ENSDART00000191606
|
CU468723.2
|
|
| chr25_+_35134393 | 0.26 |
ENSDART00000185379
|
CR762436.2
|
|
| chr7_+_5976613 | 0.26 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr25_-_7974494 | 0.26 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr21_+_19648814 | 0.26 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr7_+_34290051 | 0.26 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr9_+_2522797 | 0.25 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr17_-_26867725 | 0.25 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr16_-_30564319 | 0.25 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr24_-_21258945 | 0.25 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr12_-_23658888 | 0.25 |
ENSDART00000088319
|
map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr4_+_7508316 | 0.25 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr14_+_17382803 | 0.24 |
ENSDART00000040383
|
si:ch211-255i20.3
|
si:ch211-255i20.3 |
| chr23_-_33709964 | 0.23 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr4_-_20118468 | 0.23 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr10_+_19525839 | 0.23 |
ENSDART00000162912
ENSDART00000158512 |
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr22_-_11626014 | 0.23 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr18_+_23249519 | 0.22 |
ENSDART00000005740
ENSDART00000147446 ENSDART00000124818 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr4_+_16725960 | 0.22 |
ENSDART00000034441
|
tcp11l2
|
t-complex 11, testis-specific-like 2 |
| chr3_+_24275597 | 0.22 |
ENSDART00000183926
ENSDART00000169765 |
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr14_-_41556720 | 0.22 |
ENSDART00000149244
|
itga6l
|
integrin, alpha 6, like |
| chr22_+_15336752 | 0.22 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr14_+_36738069 | 0.22 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr7_+_29863548 | 0.21 |
ENSDART00000136555
ENSDART00000134068 |
tln2a
|
talin 2a |
| chr10_+_13209580 | 0.21 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr8_-_40555340 | 0.20 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr1_-_17570013 | 0.20 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr12_-_20665164 | 0.20 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr19_-_3381422 | 0.19 |
ENSDART00000105146
|
edn1
|
endothelin 1 |
| chr23_-_23401305 | 0.19 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr16_+_13424534 | 0.19 |
ENSDART00000114944
|
zgc:101640
|
zgc:101640 |
| chr3_+_24275766 | 0.19 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr5_-_66301142 | 0.19 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr3_+_40809892 | 0.19 |
ENSDART00000190803
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr14_-_25949951 | 0.18 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr8_+_20393250 | 0.18 |
ENSDART00000015038
|
zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr15_-_2903475 | 0.18 |
ENSDART00000043226
|
guca1c
|
guanylate cyclase activator 1C |
| chr14_-_28052474 | 0.17 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr6_+_8176486 | 0.17 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr6_+_51932563 | 0.17 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gata1b+gata2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.3 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 1.4 | 8.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.7 | 2.0 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.5 | 1.5 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.5 | 1.5 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.5 | 1.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.4 | 2.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 1.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 4.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 0.7 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.2 | 0.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 3.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.7 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 0.9 | GO:0001955 | blood vessel maturation(GO:0001955) regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 1.5 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.1 | 1.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.7 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.7 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.5 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.4 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.3 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 1.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 1.5 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 0.3 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.3 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 1.8 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.2 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 1.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 1.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.2 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 1.9 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.6 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 3.2 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 4.1 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 1.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.6 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.0 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 3.8 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 2.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 4.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 9.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 3.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.1 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 1.5 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.3 | 3.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 1.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 3.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.8 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 4.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 2.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 1.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 0.7 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 1.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.6 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 5.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0030250 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 3.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.1 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 7.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |