Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
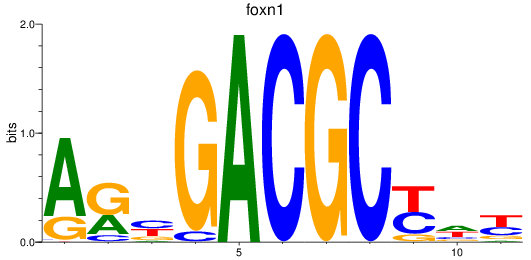
Results for foxn1
Z-value: 1.77
Transcription factors associated with foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxn1
|
ENSDARG00000011879 | forkhead box N1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxn1 | dr11_v1_chr15_-_28161224_28161224 | -0.40 | 5.1e-01 | Click! |
Activity profile of foxn1 motif
Sorted Z-values of foxn1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_21832441 | 1.72 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr13_-_51247529 | 1.26 |
ENSDART00000191774
ENSDART00000083788 |
AL929217.1
|
|
| chr5_-_31901468 | 1.22 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr7_+_23907692 | 1.16 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr23_+_19590006 | 1.13 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr12_-_19103490 | 1.06 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr17_+_29345606 | 1.06 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr25_+_12640211 | 1.06 |
ENSDART00000165108
|
jph3
|
junctophilin 3 |
| chr1_+_19930520 | 1.00 |
ENSDART00000158344
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr3_-_35602233 | 0.99 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr3_+_33340939 | 0.96 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr19_-_27966526 | 0.95 |
ENSDART00000141896
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr17_-_46457622 | 0.93 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr7_+_23495986 | 0.93 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr1_+_41690402 | 0.88 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr21_+_3093419 | 0.88 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr24_-_38374744 | 0.87 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr21_-_14251306 | 0.86 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr15_-_22147860 | 0.84 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr4_-_8152746 | 0.83 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr19_-_27966780 | 0.82 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr14_+_8275115 | 0.82 |
ENSDART00000129055
|
nrg2b
|
neuregulin 2b |
| chr8_+_6967108 | 0.81 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr4_+_2619132 | 0.80 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr12_+_26467847 | 0.80 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr11_+_6819050 | 0.79 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr13_-_50672341 | 0.79 |
ENSDART00000190498
|
FP102122.1
|
|
| chr18_-_39787040 | 0.79 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr10_+_37500234 | 0.77 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr6_+_51932267 | 0.77 |
ENSDART00000156256
|
angpt4
|
angiopoietin 4 |
| chr15_-_15449929 | 0.74 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr17_-_7861219 | 0.73 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr1_+_33969015 | 0.73 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr10_+_21668606 | 0.72 |
ENSDART00000185751
|
CT737184.2
|
|
| chr23_-_27345425 | 0.72 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr7_+_69528850 | 0.72 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr21_-_43606502 | 0.72 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr11_-_38083397 | 0.68 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr12_+_9703172 | 0.68 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr8_+_668184 | 0.65 |
ENSDART00000183788
|
rnf165b
|
ring finger protein 165b |
| chr14_-_2361692 | 0.65 |
ENSDART00000167696
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.4 |
| chr11_+_23933016 | 0.64 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr23_-_10786400 | 0.63 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr8_+_36500061 | 0.63 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr4_-_12102025 | 0.63 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr16_+_50741154 | 0.63 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr17_+_44780166 | 0.62 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr2_-_6292510 | 0.62 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr6_-_42377307 | 0.62 |
ENSDART00000129302
|
emc3
|
ER membrane protein complex subunit 3 |
| chr7_+_22801465 | 0.62 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr8_+_52530889 | 0.61 |
ENSDART00000127729
ENSDART00000170360 ENSDART00000162687 |
stambpb
|
STAM binding protein b |
| chr20_+_52442870 | 0.61 |
ENSDART00000163100
|
arhgap39
|
Rho GTPase activating protein 39 |
| chr17_-_45552602 | 0.60 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr17_+_37301860 | 0.60 |
ENSDART00000181531
ENSDART00000075978 |
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr23_+_23485858 | 0.60 |
ENSDART00000114067
|
agrn
|
agrin |
| chr20_-_28884800 | 0.59 |
ENSDART00000134564
ENSDART00000132127 ENSDART00000135506 ENSDART00000075515 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr6_+_51932563 | 0.59 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
| chr13_+_2442841 | 0.59 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr15_+_19335646 | 0.59 |
ENSDART00000062569
|
acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr23_-_35347714 | 0.59 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr19_+_5134624 | 0.57 |
ENSDART00000151324
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr5_-_29643381 | 0.56 |
ENSDART00000034849
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr1_-_21483832 | 0.55 |
ENSDART00000102790
|
glrba
|
glycine receptor, beta a |
| chr18_+_328689 | 0.55 |
ENSDART00000167841
|
ss18l2
|
synovial sarcoma translocation gene on chromosome 18-like 2 |
| chr8_+_54055390 | 0.54 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr16_-_27161410 | 0.54 |
ENSDART00000177503
|
LO017771.1
|
|
| chr12_-_25150239 | 0.53 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr8_-_18582922 | 0.52 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr12_-_35988586 | 0.50 |
ENSDART00000157746
|
pde6gb
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog b |
| chr24_-_18179535 | 0.50 |
ENSDART00000186112
|
cntnap2a
|
contactin associated protein like 2a |
| chr3_+_34919810 | 0.50 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr24_-_29997145 | 0.50 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr2_+_37480669 | 0.49 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr6_+_6491013 | 0.48 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr13_+_51579851 | 0.48 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr8_+_35172594 | 0.48 |
ENSDART00000177146
|
BX897670.1
|
|
| chr16_+_23282655 | 0.47 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr19_+_5135113 | 0.47 |
ENSDART00000151310
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr2_+_6127593 | 0.46 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr12_-_4632519 | 0.46 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr16_-_7362806 | 0.46 |
ENSDART00000166776
|
foxo6a
|
forkhead box O6 a |
| chr13_+_24287093 | 0.46 |
ENSDART00000058628
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr25_-_25142387 | 0.46 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr17_+_43032529 | 0.46 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr7_-_19146925 | 0.45 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr7_-_18168493 | 0.45 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr16_-_6205790 | 0.45 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr12_-_13318944 | 0.44 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr6_+_10450000 | 0.44 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr5_-_52216170 | 0.44 |
ENSDART00000158542
ENSDART00000192981 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr15_+_15516612 | 0.44 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr8_-_24113575 | 0.44 |
ENSDART00000099692
ENSDART00000186211 |
dclre1b
|
DNA cross-link repair 1B |
| chr8_+_36503797 | 0.43 |
ENSDART00000184785
|
slc7a4
|
solute carrier family 7, member 4 |
| chr5_-_19932621 | 0.43 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr2_-_16359042 | 0.42 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr22_+_26703026 | 0.42 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr1_+_34295925 | 0.42 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr5_+_36415978 | 0.42 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr11_-_23687158 | 0.42 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr13_+_2448251 | 0.42 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr20_-_6196989 | 0.42 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr11_-_5563498 | 0.41 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr15_+_2421432 | 0.41 |
ENSDART00000193772
|
hephl1a
|
hephaestin-like 1a |
| chr7_-_47850702 | 0.41 |
ENSDART00000109511
|
si:ch211-186j3.6
|
si:ch211-186j3.6 |
| chr9_-_24046287 | 0.40 |
ENSDART00000184313
|
ackr3a
|
atypical chemokine receptor 3a |
| chr10_+_18877362 | 0.40 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr10_+_42374957 | 0.39 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr20_-_31808779 | 0.39 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr13_-_25284716 | 0.39 |
ENSDART00000039828
|
vcla
|
vinculin a |
| chr3_+_15773991 | 0.39 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr21_+_1119046 | 0.39 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr15_+_2421729 | 0.38 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr16_+_23531583 | 0.38 |
ENSDART00000146708
|
adar
|
adenosine deaminase, RNA-specific |
| chr17_+_10593398 | 0.38 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr1_+_34203817 | 0.38 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr13_+_35925490 | 0.38 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr3_-_60571218 | 0.38 |
ENSDART00000178981
|
si:ch73-366l1.5
|
si:ch73-366l1.5 |
| chr20_-_18915376 | 0.38 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr8_-_43158486 | 0.38 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr3_-_22216771 | 0.37 |
ENSDART00000130546
|
maptb
|
microtubule-associated protein tau b |
| chr5_+_11812089 | 0.36 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr23_+_17354951 | 0.36 |
ENSDART00000189467
|
SRMS
|
zgc:194282 |
| chr4_-_1497384 | 0.36 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr19_+_7173613 | 0.36 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr1_-_25144439 | 0.36 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr4_+_12013043 | 0.35 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr1_-_45041272 | 0.35 |
ENSDART00000190615
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr13_-_30645965 | 0.34 |
ENSDART00000109307
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr17_-_8656155 | 0.34 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr6_+_39493864 | 0.33 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr7_+_40083601 | 0.33 |
ENSDART00000099046
|
zgc:112356
|
zgc:112356 |
| chr7_+_31145386 | 0.32 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr25_+_2776511 | 0.32 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr8_-_25846188 | 0.31 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr11_-_37425407 | 0.31 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr9_+_34221997 | 0.31 |
ENSDART00000028617
|
mpc2
|
mitochondrial pyruvate carrier 2 |
| chr7_+_21859337 | 0.31 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr24_+_19210001 | 0.30 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr9_+_21793565 | 0.30 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr2_+_16696052 | 0.30 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr15_+_2559875 | 0.30 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr1_+_31725154 | 0.30 |
ENSDART00000112333
ENSDART00000189801 |
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr21_-_14175838 | 0.30 |
ENSDART00000111659
|
whrna
|
whirlin a |
| chr1_-_54063520 | 0.30 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr5_-_52215926 | 0.29 |
ENSDART00000163973
ENSDART00000193602 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr9_-_28399071 | 0.29 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr19_+_19512515 | 0.29 |
ENSDART00000180065
|
jazf1a
|
JAZF zinc finger 1a |
| chr1_-_6494384 | 0.29 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr12_+_49100365 | 0.29 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr2_-_56131312 | 0.29 |
ENSDART00000097755
|
jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr18_+_19990412 | 0.29 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr7_+_66884291 | 0.29 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr2_-_2096055 | 0.28 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr9_-_48296469 | 0.28 |
ENSDART00000058255
|
bbs5
|
Bardet-Biedl syndrome 5 |
| chr21_+_21279159 | 0.28 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr7_-_12869545 | 0.28 |
ENSDART00000163045
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr2_+_42871831 | 0.28 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr16_+_48460873 | 0.28 |
ENSDART00000159902
|
ext1a
|
exostosin glycosyltransferase 1a |
| chr7_+_17782436 | 0.28 |
ENSDART00000173793
ENSDART00000165110 |
si:dkey-28a3.2
|
si:dkey-28a3.2 |
| chr22_-_11614973 | 0.27 |
ENSDART00000063135
|
pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr13_+_19884631 | 0.27 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr21_-_35325466 | 0.27 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr20_-_50014936 | 0.27 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr25_-_16782394 | 0.27 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr16_+_20871021 | 0.26 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr5_+_54585431 | 0.26 |
ENSDART00000171225
|
npr2
|
natriuretic peptide receptor 2 |
| chr10_+_36695597 | 0.26 |
ENSDART00000169015
ENSDART00000171392 |
rab6a
|
RAB6A, member RAS oncogene family |
| chr23_+_27652906 | 0.25 |
ENSDART00000077931
|
wnt1
|
wingless-type MMTV integration site family, member 1 |
| chr17_-_53022822 | 0.25 |
ENSDART00000103434
|
zgc:154061
|
zgc:154061 |
| chr25_+_36405021 | 0.25 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr12_-_19151708 | 0.25 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr19_+_20163826 | 0.25 |
ENSDART00000090942
ENSDART00000134650 |
ccdc126
|
coiled-coil domain containing 126 |
| chr3_+_17951790 | 0.25 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr17_-_23895026 | 0.25 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr5_+_26075230 | 0.24 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr20_-_4738101 | 0.23 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr19_+_26681848 | 0.23 |
ENSDART00000138322
|
si:dkey-27c15.3
|
si:dkey-27c15.3 |
| chr3_-_29891456 | 0.23 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr12_-_10674606 | 0.23 |
ENSDART00000157919
|
med24
|
mediator complex subunit 24 |
| chr13_+_21779975 | 0.23 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr1_-_41982582 | 0.23 |
ENSDART00000014678
|
adra1d
|
adrenoceptor alpha 1D |
| chr11_-_19694334 | 0.23 |
ENSDART00000054735
|
SYNPR
|
si:dkey-30j16.3 |
| chr25_+_4660639 | 0.23 |
ENSDART00000130299
|
deaf1
|
DEAF1 transcription factor |
| chr2_+_44615000 | 0.22 |
ENSDART00000188826
ENSDART00000113232 |
yeats2
|
YEATS domain containing 2 |
| chr15_+_404891 | 0.22 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr7_+_66884570 | 0.22 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr1_-_45341760 | 0.22 |
ENSDART00000149183
ENSDART00000148289 ENSDART00000110390 |
zgc:101679
|
zgc:101679 |
| chr10_+_29137482 | 0.22 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr7_-_29571615 | 0.22 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr6_+_50381347 | 0.22 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr6_-_35106425 | 0.22 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr1_+_40237276 | 0.21 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr15_-_23908779 | 0.21 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr7_+_24006875 | 0.21 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr17_-_16324565 | 0.21 |
ENSDART00000030835
|
hmbox1a
|
homeobox containing 1a |
| chr8_+_18010568 | 0.21 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr8_-_49701904 | 0.21 |
ENSDART00000142115
|
frmd3
|
FERM domain containing 3 |
| chr13_-_27916439 | 0.21 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr8_+_27555314 | 0.21 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr20_+_54333774 | 0.21 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxn1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 0.8 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 0.6 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.8 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.2 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.2 | 0.6 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 0.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.4 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.5 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.5 | GO:0086011 | membrane repolarization during action potential(GO:0086011) membrane repolarization during cardiac muscle cell action potential(GO:0086013) cardiac muscle cell membrane repolarization(GO:0099622) |
| 0.1 | 0.4 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 1.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.4 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.5 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.6 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 0.3 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.6 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.4 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 0.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.3 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.7 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.8 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.1 | GO:0072019 | proximal convoluted tubule development(GO:0072019) |
| 0.0 | 1.0 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.2 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.8 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.5 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.4 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 1.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.5 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 1.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.4 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 1.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.9 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.2 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.9 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.6 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |