Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for foxm1
Z-value: 1.65
Transcription factors associated with foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxm1
|
ENSDARG00000003200 | forkhead box M1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxm1 | dr11_v1_chr4_-_5826320_5826320 | 0.77 | 1.2e-01 | Click! |
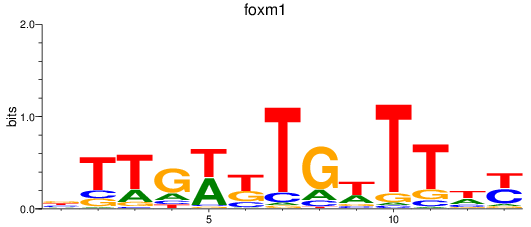
Activity profile of foxm1 motif
Sorted Z-values of foxm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_23770666 | 1.13 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr25_-_22187397 | 1.09 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr19_+_48164242 | 1.09 |
ENSDART00000186574
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr15_-_41762530 | 0.89 |
ENSDART00000187125
ENSDART00000154971 |
ftr91
|
finTRIM family, member 91 |
| chr3_-_19561058 | 0.87 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr22_+_835728 | 0.84 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr16_+_21801277 | 0.78 |
ENSDART00000088407
|
trim108
|
tripartite motif containing 108 |
| chr3_+_1167026 | 0.74 |
ENSDART00000031823
ENSDART00000155340 |
triobpb
|
TRIO and F-actin binding protein b |
| chr5_-_38197080 | 0.71 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr22_-_17671348 | 0.71 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr14_+_20893065 | 0.69 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr14_+_45559268 | 0.68 |
ENSDART00000173152
|
RARRES3
|
zgc:154040 |
| chr6_+_49881864 | 0.59 |
ENSDART00000075040
|
tubb1
|
tubulin, beta 1 class VI |
| chr7_+_15736230 | 0.57 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr3_-_33967767 | 0.56 |
ENSDART00000151493
ENSDART00000151160 |
ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr10_-_244745 | 0.56 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr20_+_16881883 | 0.55 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr3_-_49514874 | 0.54 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr5_+_37068223 | 0.54 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr4_+_15944245 | 0.53 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr14_+_901847 | 0.52 |
ENSDART00000166991
|
si:ch73-208h1.2
|
si:ch73-208h1.2 |
| chr1_+_19538299 | 0.51 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr1_+_58370526 | 0.50 |
ENSDART00000067775
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr4_+_25558849 | 0.49 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr1_+_59088205 | 0.49 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr22_+_17606863 | 0.45 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| chr14_-_22015232 | 0.45 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr21_-_13856689 | 0.45 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr18_-_16795262 | 0.44 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr16_-_25841084 | 0.44 |
ENSDART00000159217
|
CR391937.1
|
|
| chr5_-_64883082 | 0.44 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr18_-_20458412 | 0.43 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr4_-_43731342 | 0.43 |
ENSDART00000146627
|
si:ch211-226o13.3
|
si:ch211-226o13.3 |
| chr8_+_39724138 | 0.42 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr21_-_3796461 | 0.42 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr6_+_56147812 | 0.41 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr18_-_20458840 | 0.41 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr4_+_14981854 | 0.40 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr17_+_8799661 | 0.40 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr17_+_5351922 | 0.40 |
ENSDART00000105394
|
runx2a
|
runt-related transcription factor 2a |
| chr15_-_44052927 | 0.40 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr24_+_17007407 | 0.39 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr2_-_44777592 | 0.39 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr2_-_29996036 | 0.38 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr1_+_55535827 | 0.38 |
ENSDART00000152784
|
adgre16
|
adhesion G protein-coupled receptor E16 |
| chr25_+_28679672 | 0.38 |
ENSDART00000139965
ENSDART00000134072 |
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr22_-_22301672 | 0.37 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr11_+_45153104 | 0.37 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr22_+_34615591 | 0.37 |
ENSDART00000189563
|
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr17_-_53440284 | 0.37 |
ENSDART00000126976
|
mycbp
|
c-myc binding protein |
| chr10_-_42237304 | 0.37 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr19_-_340641 | 0.37 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr14_-_4177311 | 0.37 |
ENSDART00000128129
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr9_+_33216945 | 0.37 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr25_+_5039050 | 0.36 |
ENSDART00000154700
|
parvb
|
parvin, beta |
| chr16_-_24194587 | 0.36 |
ENSDART00000181520
|
rps19
|
ribosomal protein S19 |
| chr3_+_37112693 | 0.35 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr5_+_29820266 | 0.35 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr5_-_24333684 | 0.35 |
ENSDART00000051553
|
znf703
|
zinc finger protein 703 |
| chr11_-_18017918 | 0.35 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr18_+_27000850 | 0.34 |
ENSDART00000188906
ENSDART00000086131 |
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr16_-_31675669 | 0.34 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr13_+_31550185 | 0.33 |
ENSDART00000127843
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr15_+_47362728 | 0.33 |
ENSDART00000180712
|
CABZ01099833.1
|
|
| chr14_-_52480661 | 0.32 |
ENSDART00000158353
|
exosc3
|
exosome component 3 |
| chr1_-_56208151 | 0.32 |
ENSDART00000183325
|
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr20_+_6663544 | 0.32 |
ENSDART00000135390
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr7_+_10592152 | 0.32 |
ENSDART00000182624
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr11_+_36683859 | 0.32 |
ENSDART00000170102
|
si:ch211-11c3.12
|
si:ch211-11c3.12 |
| chr17_-_53439866 | 0.32 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr19_-_18626952 | 0.32 |
ENSDART00000168004
ENSDART00000162034 ENSDART00000165486 ENSDART00000167971 |
rps18
|
ribosomal protein S18 |
| chr23_+_25305431 | 0.32 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr11_-_25213651 | 0.32 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr15_+_37589698 | 0.30 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr19_+_31904836 | 0.29 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr14_-_16082806 | 0.29 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr7_+_60359347 | 0.29 |
ENSDART00000145201
ENSDART00000039827 |
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr12_-_9516981 | 0.29 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr1_+_12348213 | 0.29 |
ENSDART00000144920
ENSDART00000138759 ENSDART00000067082 |
clta
|
clathrin, light chain A |
| chr11_-_18107447 | 0.28 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr7_+_17933384 | 0.28 |
ENSDART00000173701
|
mta2
|
metastasis associated 1 family, member 2 |
| chr18_+_19120984 | 0.27 |
ENSDART00000141501
|
si:dkey-242h9.3
|
si:dkey-242h9.3 |
| chr3_-_54607166 | 0.27 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr9_+_21259820 | 0.27 |
ENSDART00000137024
ENSDART00000132324 |
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr14_-_8453192 | 0.27 |
ENSDART00000136947
|
eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr18_-_15932704 | 0.27 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr7_-_51102479 | 0.27 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr2_-_59231017 | 0.27 |
ENSDART00000143980
ENSDART00000184190 |
ftr29
|
finTRIM family, member 29 |
| chr14_+_21686207 | 0.26 |
ENSDART00000034438
|
ran
|
RAN, member RAS oncogene family |
| chr4_+_60492313 | 0.26 |
ENSDART00000191043
ENSDART00000191631 |
si:dkey-211i20.2
|
si:dkey-211i20.2 |
| chr19_-_46018152 | 0.25 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr22_+_30543437 | 0.25 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr5_-_9948497 | 0.25 |
ENSDART00000183196
|
tor4ab
|
torsin family 4, member Ab |
| chr15_-_37589600 | 0.25 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr1_-_55166511 | 0.25 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr22_+_31059919 | 0.25 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr2_-_10338759 | 0.25 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr2_+_25840463 | 0.24 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr12_+_17154655 | 0.24 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr2_+_33926911 | 0.24 |
ENSDART00000109849
ENSDART00000135884 |
kif2c
|
kinesin family member 2C |
| chr4_+_33871518 | 0.24 |
ENSDART00000185054
|
si:dkey-28i19.3
|
si:dkey-28i19.3 |
| chr9_-_31108285 | 0.24 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr18_+_27001115 | 0.24 |
ENSDART00000133547
|
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr10_+_6121558 | 0.24 |
ENSDART00000166799
ENSDART00000157947 |
tln1
|
talin 1 |
| chr17_+_23975762 | 0.24 |
ENSDART00000155941
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr9_-_38036984 | 0.24 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr4_-_16824556 | 0.24 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr22_+_1556948 | 0.24 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr11_+_45286911 | 0.23 |
ENSDART00000181763
|
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr22_+_9287929 | 0.23 |
ENSDART00000193522
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr10_-_35257458 | 0.23 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr17_-_11439815 | 0.23 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr10_-_35051691 | 0.23 |
ENSDART00000108670
ENSDART00000190711 |
supt20
|
SPT20 homolog, SAGA complex component |
| chr4_-_68913650 | 0.23 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr3_+_1107102 | 0.23 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr17_-_50301313 | 0.23 |
ENSDART00000125772
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr16_-_32303835 | 0.23 |
ENSDART00000191408
|
mms22l
|
MMS22-like, DNA repair protein |
| chr13_-_22961605 | 0.22 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr10_+_40629616 | 0.22 |
ENSDART00000147476
|
CR396590.9
|
|
| chr21_+_3796620 | 0.22 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr7_-_3639171 | 0.22 |
ENSDART00000064280
|
si:ch211-282j17.1
|
si:ch211-282j17.1 |
| chr4_-_16824231 | 0.22 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr24_+_5893134 | 0.22 |
ENSDART00000077941
|
mastl
|
microtubule associated serine/threonine kinase-like |
| chr13_+_37653851 | 0.22 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr19_-_11966015 | 0.22 |
ENSDART00000123409
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr15_-_28082310 | 0.21 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr4_-_12718367 | 0.21 |
ENSDART00000035259
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr4_+_47445318 | 0.21 |
ENSDART00000183819
|
si:dkey-124l13.1
|
si:dkey-124l13.1 |
| chr22_-_23706771 | 0.21 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr4_+_75537772 | 0.21 |
ENSDART00000168380
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr16_-_17175731 | 0.20 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr15_-_14210382 | 0.20 |
ENSDART00000179599
|
CR925813.1
|
|
| chr10_-_35052198 | 0.20 |
ENSDART00000147805
|
supt20
|
SPT20 homolog, SAGA complex component |
| chr22_+_5851122 | 0.20 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr13_+_25421531 | 0.20 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr11_-_28614608 | 0.20 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr13_+_5978809 | 0.20 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr11_-_11336986 | 0.20 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr12_+_19191787 | 0.19 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr12_-_23365737 | 0.19 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr6_-_16804001 | 0.19 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr7_+_24114694 | 0.19 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr11_-_2250767 | 0.18 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr2_+_11205795 | 0.18 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr22_+_9239831 | 0.18 |
ENSDART00000133720
|
si:ch211-250k18.5
|
si:ch211-250k18.5 |
| chr7_+_55950229 | 0.18 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr5_-_16475682 | 0.18 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr20_-_10487951 | 0.18 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr14_+_31865099 | 0.18 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr18_+_44768829 | 0.18 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr7_+_4625915 | 0.17 |
ENSDART00000108557
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr11_+_6422374 | 0.17 |
ENSDART00000183148
|
FO681393.1
|
|
| chr13_+_33655404 | 0.17 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr7_-_6444011 | 0.17 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr4_+_5196469 | 0.17 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr11_+_2416064 | 0.17 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr4_-_22472653 | 0.16 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr21_-_23046606 | 0.16 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr11_-_13611132 | 0.16 |
ENSDART00000113055
|
si:ch211-1a19.2
|
si:ch211-1a19.2 |
| chr21_+_40685895 | 0.16 |
ENSDART00000017709
|
ccdc82
|
coiled-coil domain containing 82 |
| chr7_+_4572727 | 0.16 |
ENSDART00000137830
|
si:dkey-83f18.7
|
si:dkey-83f18.7 |
| chr20_-_5291012 | 0.16 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr8_+_23147218 | 0.16 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr3_-_37082618 | 0.16 |
ENSDART00000026701
ENSDART00000110716 |
tubg1
|
tubulin, gamma 1 |
| chr12_+_30234209 | 0.16 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr11_-_18017287 | 0.16 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr13_+_44857087 | 0.16 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr6_-_16948040 | 0.16 |
ENSDART00000156433
|
pimr30
|
Pim proto-oncogene, serine/threonine kinase, related 30 |
| chr9_+_27354414 | 0.16 |
ENSDART00000163804
|
tlr20.2
|
toll-like receptor 20, tandem duplicate 2 |
| chr9_+_1137430 | 0.15 |
ENSDART00000187062
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr7_+_1505507 | 0.15 |
ENSDART00000161015
|
nop10
|
NOP10 ribonucleoprotein homolog (yeast) |
| chr21_+_37436907 | 0.15 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr5_-_67757188 | 0.15 |
ENSDART00000167168
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr4_-_17725008 | 0.15 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr4_+_59234719 | 0.15 |
ENSDART00000170724
ENSDART00000192143 ENSDART00000109914 |
znf1086
|
zinc finger protein 1086 |
| chr6_-_54444929 | 0.15 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr13_+_29470442 | 0.15 |
ENSDART00000028417
|
lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr22_-_6066867 | 0.15 |
ENSDART00000142383
|
si:dkey-19a16.1
|
si:dkey-19a16.1 |
| chr12_-_43428542 | 0.15 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr16_-_31445781 | 0.15 |
ENSDART00000056551
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr20_+_25904199 | 0.15 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr3_+_2719243 | 0.14 |
ENSDART00000189256
|
CR388047.3
|
|
| chr4_+_75577480 | 0.14 |
ENSDART00000188196
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr25_-_24202576 | 0.14 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr6_+_40922572 | 0.14 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr13_+_24679674 | 0.14 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr1_-_18585046 | 0.14 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr14_-_38826739 | 0.14 |
ENSDART00000187633
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr15_-_18232712 | 0.14 |
ENSDART00000081199
|
wu:fj20b03
|
wu:fj20b03 |
| chr16_-_53800047 | 0.14 |
ENSDART00000158047
|
znrf2b
|
zinc and ring finger 2b |
| chr3_-_11040575 | 0.13 |
ENSDART00000159802
|
CR382337.2
|
|
| chr4_+_47789703 | 0.13 |
ENSDART00000161323
|
si:ch211-196h24.2
|
si:ch211-196h24.2 |
| chr16_+_40131473 | 0.13 |
ENSDART00000155421
ENSDART00000134732 ENSDART00000138699 |
cenpw
si:ch211-195p4.4
|
centromere protein W si:ch211-195p4.4 |
| chr7_+_24115082 | 0.13 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr6_-_29288155 | 0.13 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr2_-_14793343 | 0.13 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr22_-_8692305 | 0.13 |
ENSDART00000181602
|
CR450686.4
|
|
| chr4_-_36032177 | 0.13 |
ENSDART00000170143
|
zgc:174180
|
zgc:174180 |
| chr4_-_890220 | 0.13 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr18_+_17534627 | 0.13 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 0.5 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.8 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.5 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.3 | GO:0045601 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.2 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.7 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 0.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.3 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.2 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.2 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.3 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.4 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.1 | GO:1903430 | branchiomotor neuron axon guidance(GO:0021785) negative regulation of cell maturation(GO:1903430) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0030132 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.6 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.4 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.2 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.2 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |