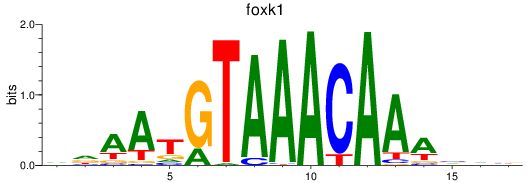
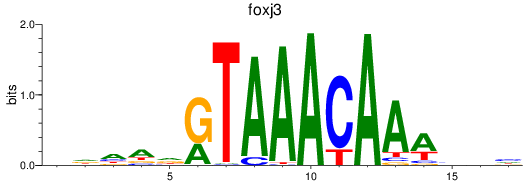
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for foxk1_foxj3
Z-value: 0.49
Transcription factors associated with foxk1_foxj3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxk1
|
ENSDARG00000037872 | forkhead box K1 |
|
foxj3
|
ENSDARG00000075774 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxk1 | dr11_v1_chr3_-_40933415_40933415 | 0.70 | 1.9e-01 | Click! |
| foxj3 | dr11_v1_chr11_-_26362294_26362334 | 0.58 | 3.0e-01 | Click! |
Activity profile of foxk1_foxj3 motif
Sorted Z-values of foxk1_foxj3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_29689981 | 0.27 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr7_-_24828296 | 0.24 |
ENSDART00000138193
|
otub1b
|
OTU deubiquitinase, ubiquitin aldehyde binding 1b |
| chr14_-_33095917 | 0.23 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr17_+_24111392 | 0.23 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr22_-_29689485 | 0.23 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr4_+_7391110 | 0.22 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr14_-_33177935 | 0.22 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr7_+_20110336 | 0.20 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr14_-_34044369 | 0.20 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr4_+_7391400 | 0.20 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr19_-_32600823 | 0.20 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr5_+_49744713 | 0.19 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr10_+_39084354 | 0.18 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr6_+_51932267 | 0.18 |
ENSDART00000156256
|
angpt4
|
angiopoietin 4 |
| chr11_+_25257022 | 0.18 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr9_-_33785093 | 0.18 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr12_+_25640480 | 0.18 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr11_+_6819050 | 0.17 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr1_+_45969240 | 0.17 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr23_-_17450746 | 0.16 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr13_+_22119798 | 0.16 |
ENSDART00000173206
ENSDART00000078652 ENSDART00000165842 |
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr24_+_17007407 | 0.15 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr6_+_51932563 | 0.15 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
| chr1_+_16127825 | 0.15 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr3_-_28258462 | 0.15 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr2_+_16696052 | 0.14 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr7_+_22849657 | 0.14 |
ENSDART00000173483
|
pygmb
|
phosphorylase, glycogen, muscle b |
| chr5_-_20446082 | 0.14 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr12_-_11457625 | 0.14 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr11_+_23933016 | 0.13 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr18_+_26899316 | 0.13 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr5_-_40190949 | 0.12 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr17_+_24718272 | 0.12 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr15_+_8043751 | 0.12 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr7_-_44963154 | 0.12 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr17_-_14966384 | 0.11 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr16_+_28578352 | 0.11 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr19_-_32600638 | 0.11 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr1_+_53945934 | 0.11 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr16_-_41131578 | 0.11 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr9_+_41459759 | 0.11 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr11_-_16152400 | 0.10 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr11_+_16153207 | 0.10 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr2_-_11662851 | 0.10 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr3_-_25369557 | 0.10 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr12_+_19188542 | 0.10 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr17_+_14965570 | 0.10 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr8_+_23093155 | 0.10 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr6_-_30839763 | 0.10 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr7_-_58098814 | 0.10 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr25_-_25142387 | 0.09 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr5_-_51998708 | 0.09 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr11_+_16152316 | 0.09 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr5_-_38384289 | 0.09 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr12_-_32066469 | 0.09 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr25_-_35360096 | 0.09 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr19_+_46222428 | 0.09 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr15_+_19990068 | 0.09 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr19_-_5103313 | 0.09 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr16_+_25011994 | 0.09 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr18_+_17428506 | 0.08 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr19_+_10396042 | 0.08 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr10_+_20070178 | 0.08 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr15_+_20543770 | 0.08 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr7_-_7398350 | 0.08 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr11_-_12051805 | 0.08 |
ENSDART00000110117
ENSDART00000182744 |
socs7
|
suppressor of cytokine signaling 7 |
| chr15_+_37589698 | 0.08 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr22_+_26798853 | 0.08 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr2_+_27330461 | 0.08 |
ENSDART00000087643
|
tesk2
|
testis-specific kinase 2 |
| chr19_+_46222918 | 0.08 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr10_-_14929630 | 0.08 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr1_-_9940494 | 0.08 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr16_+_6944564 | 0.08 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr5_+_36611128 | 0.07 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr5_+_61799629 | 0.07 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr18_+_6857071 | 0.07 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr13_+_22280983 | 0.07 |
ENSDART00000173258
ENSDART00000173379 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr1_+_25801648 | 0.07 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr1_-_30689004 | 0.07 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr16_+_10413551 | 0.07 |
ENSDART00000032877
ENSDART00000172827 |
ino80e
|
INO80 complex subunit E |
| chr24_-_21588914 | 0.07 |
ENSDART00000152054
|
gpr12
|
G protein-coupled receptor 12 |
| chr19_-_47997424 | 0.07 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr3_-_26191960 | 0.07 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr13_+_30951155 | 0.07 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr24_-_25004553 | 0.07 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr3_-_60142530 | 0.07 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr16_-_43025885 | 0.07 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr21_+_28502340 | 0.07 |
ENSDART00000077897
ENSDART00000140229 |
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr12_-_29624638 | 0.07 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr16_+_6944311 | 0.07 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr13_+_9368621 | 0.07 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr16_-_26820634 | 0.07 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr3_+_27786601 | 0.06 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr8_+_16990120 | 0.06 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr23_-_32304650 | 0.06 |
ENSDART00000143772
ENSDART00000085642 ENSDART00000188989 |
dgkaa
|
diacylglycerol kinase, alpha a |
| chr9_+_38163876 | 0.06 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr1_-_12394048 | 0.06 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr8_-_39859688 | 0.06 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr19_+_20177887 | 0.06 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr19_+_2275019 | 0.06 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr2_-_8017579 | 0.06 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr22_+_4707663 | 0.06 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr2_-_30200206 | 0.06 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr5_+_12528693 | 0.06 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr5_-_45894802 | 0.06 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr13_-_34781984 | 0.06 |
ENSDART00000172138
|
ism1
|
isthmin 1 |
| chr3_+_19685873 | 0.06 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr20_-_19365875 | 0.06 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr13_-_21701323 | 0.06 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr8_-_1267247 | 0.06 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr1_-_9277986 | 0.06 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr21_-_27272657 | 0.06 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr19_-_22843480 | 0.06 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr19_-_5103141 | 0.06 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_-_28209001 | 0.06 |
ENSDART00000151178
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr16_+_31827502 | 0.06 |
ENSDART00000045210
|
mlf2
|
myeloid leukemia factor 2 |
| chr18_-_13121983 | 0.06 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr21_-_28920245 | 0.06 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr4_-_20135406 | 0.06 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr3_+_46762703 | 0.06 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr11_-_183328 | 0.06 |
ENSDART00000168562
|
avil
|
advillin |
| chr5_-_19963537 | 0.05 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr3_+_34821327 | 0.05 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr21_-_13661631 | 0.05 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr2_+_1988036 | 0.05 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr11_-_13341483 | 0.05 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr4_-_75157223 | 0.05 |
ENSDART00000174127
|
CABZ01066312.1
|
|
| chr5_+_24305877 | 0.05 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr21_+_19547806 | 0.05 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr21_+_25071805 | 0.05 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr7_-_41013575 | 0.05 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr5_-_66749535 | 0.05 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr1_+_50538839 | 0.05 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr16_-_15263099 | 0.05 |
ENSDART00000125691
|
sntb1
|
syntrophin, basic 1 |
| chr2_+_5563077 | 0.05 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr16_+_31827653 | 0.05 |
ENSDART00000187261
|
mlf2
|
myeloid leukemia factor 2 |
| chr5_+_15495351 | 0.05 |
ENSDART00000111646
ENSDART00000114446 |
suds3
|
SDS3 homolog, SIN3A corepressor complex component |
| chr11_+_2710530 | 0.05 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr11_-_13341051 | 0.05 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr19_-_28367413 | 0.05 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr21_-_27273147 | 0.05 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr10_-_14929392 | 0.05 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr20_-_19590378 | 0.05 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr24_+_11381400 | 0.05 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr14_-_48939560 | 0.05 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr25_+_17920361 | 0.05 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr18_+_20226843 | 0.05 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr3_+_19207176 | 0.05 |
ENSDART00000087803
|
rln3a
|
relaxin 3a |
| chr13_+_5978809 | 0.05 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr23_+_7692042 | 0.05 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr20_-_26039841 | 0.04 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr17_-_35352690 | 0.04 |
ENSDART00000016053
|
rnf144aa
|
ring finger protein 144aa |
| chr15_+_30157602 | 0.04 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr18_-_6855991 | 0.04 |
ENSDART00000135206
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr2_+_26179096 | 0.04 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr15_+_23784842 | 0.04 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr17_+_32500387 | 0.04 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr25_+_17920668 | 0.04 |
ENSDART00000093358
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr14_-_26177156 | 0.04 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr4_-_22472653 | 0.04 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr21_-_27881752 | 0.04 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr19_+_5315987 | 0.04 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr8_-_7567815 | 0.04 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr20_-_18736281 | 0.04 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr23_-_27608257 | 0.04 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr2_-_24348642 | 0.04 |
ENSDART00000181739
|
ano8a
|
anoctamin 8a |
| chr11_-_21586157 | 0.04 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr3_+_55031685 | 0.04 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr13_+_31108334 | 0.04 |
ENSDART00000142245
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr23_+_31596441 | 0.04 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr25_-_25434479 | 0.04 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr2_-_24444838 | 0.04 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr16_-_17162843 | 0.04 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr16_-_5105295 | 0.04 |
ENSDART00000082071
ENSDART00000148955 ENSDART00000184700 ENSDART00000188127 |
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr23_-_16682186 | 0.04 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr5_-_12743196 | 0.04 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr22_-_18179214 | 0.04 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr11_+_11120532 | 0.04 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr14_-_41478265 | 0.04 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr15_+_39977461 | 0.04 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr6_-_8466717 | 0.04 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr5_+_61361815 | 0.04 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr4_-_20135919 | 0.04 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr25_-_18953322 | 0.04 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr10_-_36633882 | 0.04 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr24_-_2312868 | 0.04 |
ENSDART00000140125
ENSDART00000138432 |
cul2
|
cullin 2 |
| chr13_+_38430466 | 0.04 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr16_+_9580699 | 0.04 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr23_+_33987382 | 0.04 |
ENSDART00000037501
|
fam50a
|
family with sequence similarity 50, member A |
| chr6_-_14038804 | 0.03 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr10_-_41156348 | 0.03 |
ENSDART00000058622
|
aak1b
|
AP2 associated kinase 1b |
| chr16_+_43152727 | 0.03 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr2_-_24603325 | 0.03 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr23_+_31596194 | 0.03 |
ENSDART00000160748
|
tbpl1
|
TBP-like 1 |
| chr20_-_35470891 | 0.03 |
ENSDART00000152993
ENSDART00000016090 |
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr5_-_56924747 | 0.03 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr6_-_10305918 | 0.03 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr13_-_25842074 | 0.03 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr12_+_5081759 | 0.03 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr18_+_21113216 | 0.03 |
ENSDART00000060191
|
kmt5b
|
lysine methyltransferase 5B |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxk1_foxj3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.5 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.1 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.0 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.0 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0045191 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.0 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |