Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
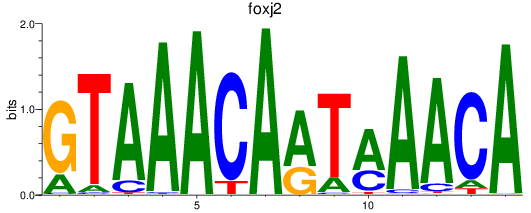
Results for foxj2
Z-value: 0.81
Transcription factors associated with foxj2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj2
|
ENSDARG00000057680 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj2 | dr11_v1_chr16_-_12784373_12784373 | 0.59 | 2.9e-01 | Click! |
Activity profile of foxj2 motif
Sorted Z-values of foxj2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48340774 | 0.71 |
ENSDART00000168065
|
si:ch73-359m17.5
|
si:ch73-359m17.5 |
| chr4_+_76671012 | 0.69 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr16_+_44768361 | 0.68 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr2_-_36918709 | 0.66 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr5_-_63302944 | 0.65 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr15_-_29598444 | 0.65 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr1_-_45177373 | 0.62 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr15_+_42573909 | 0.57 |
ENSDART00000181801
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr4_-_77680276 | 0.48 |
ENSDART00000153262
|
si:ch211-250m6.2
|
si:ch211-250m6.2 |
| chr8_+_32406885 | 0.41 |
ENSDART00000167600
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr18_+_7553950 | 0.38 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr19_-_2318391 | 0.38 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr20_+_34770197 | 0.37 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr18_-_44316920 | 0.37 |
ENSDART00000098599
|
si:ch211-151h10.2
|
si:ch211-151h10.2 |
| chr11_+_37768298 | 0.37 |
ENSDART00000166886
|
sox13
|
SRY (sex determining region Y)-box 13 |
| chr4_-_12795436 | 0.37 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr19_+_18739085 | 0.36 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr19_+_6990970 | 0.31 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr18_-_15911394 | 0.30 |
ENSDART00000091339
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr9_-_6380653 | 0.30 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr4_-_12795030 | 0.29 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr13_-_2010191 | 0.29 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr16_+_35916371 | 0.29 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr15_-_29162193 | 0.28 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr8_-_39838660 | 0.26 |
ENSDART00000139266
|
ftr98
|
finTRIM family, member 98 |
| chr6_-_14004772 | 0.26 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr5_-_69212184 | 0.26 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr14_+_21699414 | 0.25 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr23_-_17450746 | 0.25 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr18_+_35742838 | 0.24 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr25_-_19608382 | 0.24 |
ENSDART00000022279
ENSDART00000135201 ENSDART00000147223 ENSDART00000190220 ENSDART00000184242 ENSDART00000166824 |
gtse1
|
G-2 and S-phase expressed 1 |
| chr13_-_37647209 | 0.23 |
ENSDART00000189102
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr14_+_21699129 | 0.23 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr13_-_15986871 | 0.23 |
ENSDART00000189394
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr10_-_35257458 | 0.21 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr10_-_15672862 | 0.21 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr24_+_38201089 | 0.20 |
ENSDART00000132338
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr16_+_33143503 | 0.20 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr1_-_55166511 | 0.19 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr2_+_39618951 | 0.19 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr8_-_46457233 | 0.18 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr1_+_17527931 | 0.18 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr18_+_20034023 | 0.18 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr1_+_9994811 | 0.17 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr8_+_20918207 | 0.16 |
ENSDART00000144039
|
si:ch73-196i15.5
|
si:ch73-196i15.5 |
| chr15_-_5901514 | 0.16 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr11_-_11878099 | 0.16 |
ENSDART00000188314
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr5_+_22970617 | 0.15 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr9_-_34842414 | 0.15 |
ENSDART00000126348
|
BX601644.1
|
Danio rerio cytokine receptor-like factor 2 (crlf2), mRNA. |
| chr25_+_3217419 | 0.15 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr2_-_29996036 | 0.15 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr17_+_5985933 | 0.15 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr17_+_51743908 | 0.15 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr6_-_7439490 | 0.15 |
ENSDART00000188825
|
fkbp11
|
FK506 binding protein 11 |
| chr15_-_9002155 | 0.14 |
ENSDART00000126708
|
rhoub
|
ras homolog family member Ub |
| chr17_+_33415319 | 0.14 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr17_-_53439866 | 0.14 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr12_-_17479078 | 0.14 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr14_-_5690363 | 0.14 |
ENSDART00000144539
|
tlx2
|
T cell leukemia homeobox 2 |
| chr14_+_20911310 | 0.13 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr16_+_10422836 | 0.13 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr7_-_54679595 | 0.13 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr3_-_57762247 | 0.13 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr6_-_18393476 | 0.13 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr21_+_45757317 | 0.12 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr1_-_9940494 | 0.12 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr19_+_20177887 | 0.12 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr3_-_14571514 | 0.12 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr10_-_1180314 | 0.12 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr6_+_7466223 | 0.11 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr2_-_39017838 | 0.11 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr17_+_33415542 | 0.11 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr13_-_869808 | 0.11 |
ENSDART00000189029
|
AL929536.6
|
|
| chr25_+_17920361 | 0.10 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr3_+_59864872 | 0.10 |
ENSDART00000102014
|
mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr15_+_21254800 | 0.10 |
ENSDART00000142902
|
usf1
|
upstream transcription factor 1 |
| chr19_+_46222428 | 0.10 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr25_+_16689633 | 0.10 |
ENSDART00000073416
|
ada2a
|
adenosine deaminase 2a |
| chr3_+_32112004 | 0.09 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr19_-_11846958 | 0.09 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr25_-_35182347 | 0.09 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr15_+_11883804 | 0.09 |
ENSDART00000163255
|
gpr184
|
G protein-coupled receptor 184 |
| chr19_+_48018802 | 0.09 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr14_-_34633960 | 0.09 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr25_-_3217115 | 0.08 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr6_+_12527725 | 0.08 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr4_+_14727018 | 0.08 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr18_+_38885309 | 0.08 |
ENSDART00000041597
|
arpp19a
|
cAMP-regulated phosphoprotein 19a |
| chr17_-_28797395 | 0.08 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr4_-_16353733 | 0.08 |
ENSDART00000186785
|
lum
|
lumican |
| chr16_+_54210554 | 0.08 |
ENSDART00000172622
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr16_+_45922175 | 0.08 |
ENSDART00000018253
|
rbm8a
|
RNA binding motif protein 8A |
| chr3_+_32129632 | 0.08 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr11_+_3575963 | 0.08 |
ENSDART00000077305
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr23_-_10914275 | 0.08 |
ENSDART00000112965
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr2_+_22602301 | 0.08 |
ENSDART00000038514
|
sept2
|
septin 2 |
| chr16_-_5154024 | 0.07 |
ENSDART00000132069
ENSDART00000060635 |
dctn3
|
dynactin 3 (p22) |
| chr9_-_32158288 | 0.07 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr22_-_9728208 | 0.07 |
ENSDART00000185962
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr2_+_29996650 | 0.07 |
ENSDART00000138050
ENSDART00000141026 |
rbm33b
|
RNA binding motif protein 33b |
| chr8_+_10305400 | 0.07 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr4_+_43700319 | 0.07 |
ENSDART00000141967
|
si:ch211-226o13.1
|
si:ch211-226o13.1 |
| chr14_+_6991142 | 0.07 |
ENSDART00000157635
ENSDART00000059918 |
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr1_-_49947290 | 0.07 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr10_+_36029537 | 0.07 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr23_+_14217508 | 0.07 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr11_-_1400507 | 0.07 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr24_+_17345521 | 0.06 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr3_-_58046319 | 0.06 |
ENSDART00000155968
|
si:ch211-256e16.10
|
si:ch211-256e16.10 |
| chr22_+_4442473 | 0.06 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr6_-_29288155 | 0.06 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr10_-_11261565 | 0.06 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr15_-_36357889 | 0.06 |
ENSDART00000156377
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr4_+_17353714 | 0.06 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr25_+_17920668 | 0.06 |
ENSDART00000093358
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr18_-_17721810 | 0.06 |
ENSDART00000061016
|
fam192a
|
family with sequence similarity 192, member A |
| chr19_+_19767567 | 0.06 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr4_+_14727212 | 0.06 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr1_-_26444075 | 0.06 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr4_-_31700186 | 0.05 |
ENSDART00000183986
ENSDART00000180890 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr21_+_28724099 | 0.05 |
ENSDART00000138017
|
puraa
|
purine-rich element binding protein Aa |
| chr17_+_38602790 | 0.05 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr16_-_17560822 | 0.05 |
ENSDART00000089230
|
casp2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr19_+_48018464 | 0.05 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr9_+_8942258 | 0.05 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr2_-_51507540 | 0.05 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr14_+_35279343 | 0.05 |
ENSDART00000187750
|
clint1a
|
clathrin interactor 1a |
| chr19_-_15229421 | 0.04 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr10_+_15025006 | 0.04 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr23_+_20640875 | 0.04 |
ENSDART00000147382
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr23_+_40275400 | 0.04 |
ENSDART00000184259
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr4_-_75681004 | 0.04 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr7_+_67434677 | 0.04 |
ENSDART00000165971
ENSDART00000166702 |
kars
|
lysyl-tRNA synthetase |
| chr20_+_33924235 | 0.04 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr10_+_45031398 | 0.04 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr5_+_63302660 | 0.04 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr19_+_46222918 | 0.04 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr22_-_718615 | 0.04 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr1_-_23268013 | 0.04 |
ENSDART00000146575
|
rfc1
|
replication factor C (activator 1) 1 |
| chr17_+_16755287 | 0.04 |
ENSDART00000080129
|
ston2
|
stonin 2 |
| chr9_+_11293830 | 0.04 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr5_+_12590403 | 0.04 |
ENSDART00000133167
|
si:dkey-98f17.5
|
si:dkey-98f17.5 |
| chr25_-_37465064 | 0.04 |
ENSDART00000186128
|
zgc:158366
|
zgc:158366 |
| chr12_+_5977777 | 0.04 |
ENSDART00000152302
|
si:ch211-131k2.2
|
si:ch211-131k2.2 |
| chr2_-_38337122 | 0.04 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr6_+_11829867 | 0.04 |
ENSDART00000151044
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr8_+_30709685 | 0.03 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr21_-_34261677 | 0.03 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr3_+_14571813 | 0.03 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr19_-_1920613 | 0.03 |
ENSDART00000186176
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr1_+_34527213 | 0.03 |
ENSDART00000180483
|
klf12a
|
Kruppel-like factor 12a |
| chr8_+_23147609 | 0.03 |
ENSDART00000180284
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr23_+_40275601 | 0.03 |
ENSDART00000076876
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr2_-_31376606 | 0.03 |
ENSDART00000098988
ENSDART00000125746 |
clul1
|
clusterin-like 1 (retinal) |
| chr14_-_25577094 | 0.03 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr13_-_377256 | 0.03 |
ENSDART00000137398
|
ch1073-291c23.1
|
ch1073-291c23.1 |
| chr18_-_24988645 | 0.03 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr11_-_22372072 | 0.03 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr17_-_681142 | 0.03 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr19_+_41551335 | 0.03 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr8_+_48942470 | 0.03 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr3_-_37588855 | 0.02 |
ENSDART00000149258
|
arf2a
|
ADP-ribosylation factor 2a |
| chr3_-_44094451 | 0.02 |
ENSDART00000157902
ENSDART00000190036 ENSDART00000188028 |
kdm8
|
lysine (K)-specific demethylase 8 |
| chr3_+_28860283 | 0.02 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr11_-_45420212 | 0.02 |
ENSDART00000182042
ENSDART00000163185 |
ankrd13c
|
ankyrin repeat domain 13C |
| chr18_+_7611298 | 0.02 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr10_-_13178853 | 0.02 |
ENSDART00000163740
ENSDART00000166327 ENSDART00000160265 ENSDART00000164299 |
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr17_+_15534815 | 0.02 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr17_+_11372531 | 0.02 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr7_-_20464468 | 0.02 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr23_-_7594723 | 0.02 |
ENSDART00000115298
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr16_+_2565433 | 0.02 |
ENSDART00000188014
ENSDART00000171378 |
preb
|
prolactin regulatory element binding |
| chr8_+_7097929 | 0.02 |
ENSDART00000188955
ENSDART00000184772 ENSDART00000109581 |
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr5_-_9678075 | 0.02 |
ENSDART00000097217
|
FAM109B
|
si:ch211-193c2.2 |
| chr5_+_62340799 | 0.02 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr13_-_25476920 | 0.02 |
ENSDART00000131403
|
tial1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr3_-_44094248 | 0.02 |
ENSDART00000166866
|
kdm8
|
lysine (K)-specific demethylase 8 |
| chr3_-_62194512 | 0.02 |
ENSDART00000074174
|
tbl3
|
transducin (beta)-like 3 |
| chr10_+_42898103 | 0.02 |
ENSDART00000015872
|
zcchc9
|
zinc finger, CCHC domain containing 9 |
| chr2_-_5356686 | 0.02 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr19_-_35492693 | 0.02 |
ENSDART00000135838
ENSDART00000051745 ENSDART00000177052 |
ptp4a2b
|
protein tyrosine phosphatase type IVA, member 2b |
| chr1_-_14503182 | 0.01 |
ENSDART00000190599
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr16_-_41646164 | 0.01 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr25_-_6391271 | 0.01 |
ENSDART00000138782
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr19_-_6134802 | 0.01 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr8_-_51954562 | 0.01 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
| chr20_+_34455645 | 0.01 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr17_-_39761086 | 0.01 |
ENSDART00000032410
|
gpr132a
|
G protein-coupled receptor 132a |
| chr13_+_47007075 | 0.01 |
ENSDART00000109247
ENSDART00000183205 ENSDART00000180924 ENSDART00000133146 |
anapc1
|
anaphase promoting complex subunit 1 |
| chr18_+_41561285 | 0.01 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr12_-_44018667 | 0.01 |
ENSDART00000170692
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr5_+_42957503 | 0.01 |
ENSDART00000192885
|
mob1ba
|
MOB kinase activator 1Ba |
| chr8_+_31016180 | 0.01 |
ENSDART00000130870
ENSDART00000143604 |
odf2b
|
outer dense fiber of sperm tails 2b |
| chr9_+_12887491 | 0.01 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr3_-_29891218 | 0.01 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr8_-_10949847 | 0.01 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr2_+_20410652 | 0.01 |
ENSDART00000185940
|
palmda
|
palmdelphin a |
| chr4_+_21741228 | 0.01 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr16_-_32975951 | 0.01 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr5_-_24201437 | 0.01 |
ENSDART00000114113
|
sox19a
|
SRY (sex determining region Y)-box 19a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.4 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.7 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.4 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.0 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |