Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
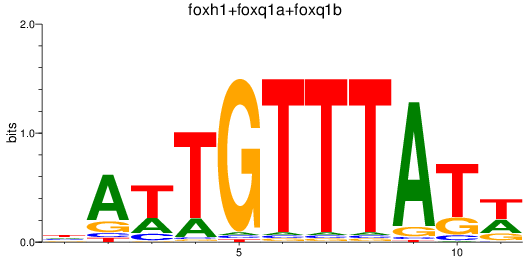
Results for foxh1+foxq1a+foxq1b
Z-value: 0.97
Transcription factors associated with foxh1+foxq1a+foxq1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxq1a
|
ENSDARG00000030896 | forkhead box Q1a |
|
foxq1b
|
ENSDARG00000055395 | forkhead box Q1b |
|
foxh1
|
ENSDARG00000055630 | forkhead box H1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxh1 | dr11_v1_chr12_-_13730501_13730501 | 0.88 | 4.6e-02 | Click! |
| foxq1a | dr11_v1_chr2_-_722156_722170 | -0.45 | 4.5e-01 | Click! |
| foxq1b | dr11_v1_chr20_+_26683933_26683940 | -0.03 | 9.6e-01 | Click! |
Activity profile of foxh1+foxq1a+foxq1b motif
Sorted Z-values of foxh1+foxq1a+foxq1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37837245 | 1.50 |
ENSDART00000171617
|
epd
|
ependymin |
| chr16_-_28856112 | 0.75 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr4_+_75200467 | 0.54 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr15_+_7992906 | 0.54 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr11_-_8126223 | 0.53 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr6_-_9792004 | 0.50 |
ENSDART00000081129
|
cdk15
|
cyclin-dependent kinase 15 |
| chr1_+_9290103 | 0.50 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr17_+_15534815 | 0.49 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr25_+_7229046 | 0.47 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr9_+_31282161 | 0.46 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr14_+_33458294 | 0.46 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr3_+_29714775 | 0.46 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr4_+_8797197 | 0.45 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr5_+_49744713 | 0.45 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr14_-_40389699 | 0.44 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr17_-_19022990 | 0.44 |
ENSDART00000154186
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr14_-_33177935 | 0.43 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr6_+_13787855 | 0.42 |
ENSDART00000182899
|
tmem198b
|
transmembrane protein 198b |
| chr8_+_39634114 | 0.41 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr2_-_44282796 | 0.38 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr1_+_44711446 | 0.38 |
ENSDART00000193481
ENSDART00000003895 |
ssrp1b
|
structure specific recognition protein 1b |
| chr21_-_32487061 | 0.38 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr1_+_53321878 | 0.38 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr3_+_34821327 | 0.38 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr2_-_44283554 | 0.37 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr16_+_31853919 | 0.37 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr14_-_21219659 | 0.36 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr5_+_19712011 | 0.36 |
ENSDART00000131924
|
fam222a
|
family with sequence similarity 222, member A |
| chr14_-_21218891 | 0.36 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr13_-_27675212 | 0.34 |
ENSDART00000141035
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr15_+_29662401 | 0.34 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr22_+_24559947 | 0.33 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr24_+_26039464 | 0.33 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr5_+_1278092 | 0.33 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr25_-_7925269 | 0.32 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr4_-_75157223 | 0.31 |
ENSDART00000174127
|
CABZ01066312.1
|
|
| chr5_-_13766651 | 0.31 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr9_+_18716485 | 0.30 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr17_-_42213285 | 0.30 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr21_+_22630627 | 0.30 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr15_+_23550890 | 0.29 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr22_-_29689981 | 0.29 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr3_-_49138004 | 0.29 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr1_+_24076243 | 0.29 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr3_-_42086577 | 0.28 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr4_+_12111154 | 0.28 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr14_-_24410673 | 0.28 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr2_+_24199073 | 0.28 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr18_+_7286788 | 0.28 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr13_-_3155243 | 0.28 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr3_-_39695856 | 0.28 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr17_-_29902187 | 0.27 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr1_-_53756851 | 0.26 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr7_+_33279108 | 0.25 |
ENSDART00000084530
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr1_+_44710955 | 0.25 |
ENSDART00000131296
ENSDART00000142187 |
ssrp1b
|
structure specific recognition protein 1b |
| chr1_+_46194333 | 0.25 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr4_-_6567355 | 0.25 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr11_+_37612214 | 0.24 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr4_-_12102025 | 0.24 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr23_+_6795531 | 0.24 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr17_+_24590177 | 0.24 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr14_+_34966598 | 0.23 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr6_-_6423885 | 0.23 |
ENSDART00000092257
|
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr11_+_7580079 | 0.23 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr7_+_28724919 | 0.23 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr19_+_5143128 | 0.23 |
ENSDART00000138011
ENSDART00000108541 |
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr3_-_39696066 | 0.23 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr8_-_13210959 | 0.23 |
ENSDART00000142224
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr9_+_20554896 | 0.22 |
ENSDART00000144248
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr10_+_42678520 | 0.22 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr11_-_27057572 | 0.22 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr23_+_6795709 | 0.22 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr24_+_39586701 | 0.21 |
ENSDART00000136447
|
si:dkey-161j23.7
|
si:dkey-161j23.7 |
| chr22_+_39054012 | 0.21 |
ENSDART00000191326
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr10_-_20669635 | 0.21 |
ENSDART00000131361
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr7_+_22801465 | 0.21 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr3_-_60142530 | 0.21 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr21_-_1799265 | 0.21 |
ENSDART00000066623
|
st8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr2_+_31957554 | 0.21 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr20_-_45062514 | 0.21 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr12_+_27704015 | 0.21 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr14_-_32403554 | 0.20 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr20_-_16156419 | 0.20 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr23_+_22819971 | 0.20 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr5_-_28767573 | 0.20 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr3_+_54047342 | 0.20 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr25_+_388258 | 0.20 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr12_+_32729470 | 0.20 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr18_-_46957840 | 0.19 |
ENSDART00000182754
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr16_+_28383758 | 0.19 |
ENSDART00000059038
ENSDART00000141061 |
itga8
|
integrin, alpha 8 |
| chr8_-_16592491 | 0.19 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr2_+_20472150 | 0.19 |
ENSDART00000168537
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr3_+_23488652 | 0.19 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr3_-_56896702 | 0.19 |
ENSDART00000023265
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr17_+_28005763 | 0.18 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr2_+_6126086 | 0.18 |
ENSDART00000179962
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr10_+_20180163 | 0.18 |
ENSDART00000080016
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr21_+_17542473 | 0.18 |
ENSDART00000005750
ENSDART00000141326 |
stom
|
stomatin |
| chr3_+_50172452 | 0.18 |
ENSDART00000191224
|
epn3a
|
epsin 3a |
| chr3_+_34234029 | 0.18 |
ENSDART00000044859
|
znf207a
|
zinc finger protein 207, a |
| chr21_-_13225402 | 0.17 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr14_+_28486213 | 0.17 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr18_-_17001056 | 0.16 |
ENSDART00000112627
|
tph2
|
tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) |
| chr17_-_44440832 | 0.16 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr3_-_43356082 | 0.16 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr12_-_5455936 | 0.16 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr12_-_26609347 | 0.16 |
ENSDART00000035925
|
kctd2
|
potassium channel tetramerization domain containing 2 |
| chr14_-_2933185 | 0.16 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr9_+_7030016 | 0.15 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr22_-_29906764 | 0.15 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr11_+_25257022 | 0.15 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr13_+_23157053 | 0.15 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr20_-_9963713 | 0.15 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr14_-_30960470 | 0.15 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr18_+_13077800 | 0.15 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr1_+_35790082 | 0.15 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr24_+_32472155 | 0.15 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr7_-_26076970 | 0.15 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr7_+_29080684 | 0.15 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr16_+_25116827 | 0.14 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr23_+_2191773 | 0.14 |
ENSDART00000190876
ENSDART00000126768 |
cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr2_+_33335911 | 0.14 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_+_24046179 | 0.14 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr24_+_11381400 | 0.14 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr12_-_34887943 | 0.14 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr24_+_7336807 | 0.14 |
ENSDART00000137010
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr10_+_43039947 | 0.14 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr13_+_39182099 | 0.14 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr10_+_31646020 | 0.14 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr14_+_34495216 | 0.14 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr12_-_31422433 | 0.13 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr1_+_40237276 | 0.13 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr8_-_11324143 | 0.13 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr19_-_17526735 | 0.13 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr1_-_45580787 | 0.13 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_+_4803906 | 0.13 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr13_+_12396316 | 0.13 |
ENSDART00000143557
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr6_+_11250033 | 0.12 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr18_+_40355408 | 0.12 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr25_-_4717462 | 0.12 |
ENSDART00000012603
|
drd4a
|
dopamine receptor D4a |
| chr12_+_22823219 | 0.12 |
ENSDART00000153199
ENSDART00000192571 |
afap1
|
actin filament associated protein 1 |
| chr25_-_3647277 | 0.12 |
ENSDART00000166363
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr17_+_24111392 | 0.11 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr19_+_26718074 | 0.11 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr3_-_41292569 | 0.11 |
ENSDART00000111856
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr16_+_46410520 | 0.11 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr18_+_14693682 | 0.11 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr5_-_33769211 | 0.11 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr3_+_40289418 | 0.11 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr10_-_10607118 | 0.10 |
ENSDART00000101089
|
dbh
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr11_+_24348425 | 0.10 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr6_+_11250316 | 0.10 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr6_+_33881372 | 0.10 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr15_+_42285643 | 0.10 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr18_+_6039141 | 0.10 |
ENSDART00000138972
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr13_-_23956178 | 0.10 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr7_+_30240791 | 0.10 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr10_+_9595575 | 0.10 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr1_+_51191049 | 0.10 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr10_+_25355308 | 0.10 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr6_+_40874206 | 0.09 |
ENSDART00000004295
|
zgc:112163
|
zgc:112163 |
| chr25_+_24291156 | 0.09 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr12_-_28537615 | 0.09 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr17_-_32698503 | 0.09 |
ENSDART00000157175
ENSDART00000077439 |
kcnk2a
|
potassium channel, subfamily K, member 2a |
| chr22_-_15437242 | 0.09 |
ENSDART00000063719
|
rpp21
|
ribonuclease P 21 subunit |
| chr19_-_4851411 | 0.09 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr18_+_16246806 | 0.09 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr15_+_32643873 | 0.09 |
ENSDART00000189433
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr7_+_24153070 | 0.09 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr2_+_8649293 | 0.09 |
ENSDART00000081442
ENSDART00000186024 |
fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr25_+_2776865 | 0.08 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr23_-_43595956 | 0.08 |
ENSDART00000162186
|
itchb
|
itchy E3 ubiquitin protein ligase b |
| chr25_+_36045072 | 0.08 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr3_-_35554809 | 0.08 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr19_+_4139065 | 0.08 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr3_+_18062871 | 0.08 |
ENSDART00000112384
|
ankrd40
|
ankyrin repeat domain 40 |
| chr15_-_17169935 | 0.08 |
ENSDART00000110111
|
cul5a
|
cullin 5a |
| chr18_-_45761868 | 0.07 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr13_-_23956361 | 0.07 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr17_-_12764360 | 0.07 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr12_+_19408373 | 0.07 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr21_-_40880317 | 0.07 |
ENSDART00000100054
ENSDART00000137696 |
elnb
|
elastin b |
| chr21_+_26733529 | 0.07 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr9_-_30494727 | 0.07 |
ENSDART00000148729
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr17_-_42218652 | 0.07 |
ENSDART00000081396
ENSDART00000190007 |
nkx2.2a
|
NK2 homeobox 2a |
| chr22_-_7461603 | 0.07 |
ENSDART00000170630
|
BX511034.6
|
|
| chr5_-_43682930 | 0.07 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr10_-_27009413 | 0.07 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr4_-_17642168 | 0.07 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr4_-_18436899 | 0.06 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr3_+_22442445 | 0.06 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr17_+_25856671 | 0.06 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr20_+_33294428 | 0.06 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr16_-_21692024 | 0.06 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr17_+_44441042 | 0.06 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr15_+_32642878 | 0.05 |
ENSDART00000159932
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr15_-_7337537 | 0.05 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr8_-_7847921 | 0.05 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr15_+_20843161 | 0.05 |
ENSDART00000153726
|
ulk2
|
unc-51 like autophagy activating kinase 2 |
| chr4_+_15011341 | 0.05 |
ENSDART00000124452
|
ube2h
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxh1+foxq1a+foxq1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.5 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.1 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.9 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.2 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.3 | GO:0099590 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.3 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.4 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.3 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |