Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
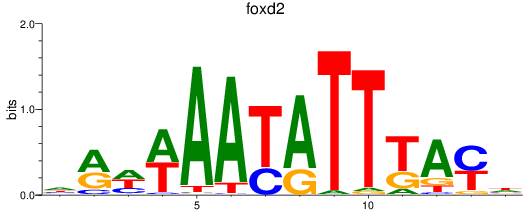
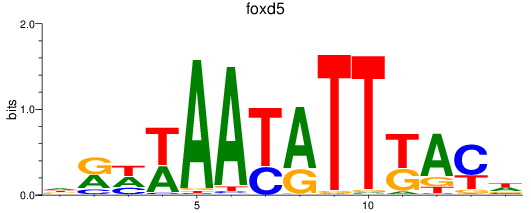
Results for foxd2_foxd5
Z-value: 1.94
Transcription factors associated with foxd2_foxd5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd2
|
ENSDARG00000058133 | forkhead box D2 |
|
foxd5
|
ENSDARG00000042485 | forkhead box D5 |
|
foxd5
|
ENSDARG00000109712 | forkhead box D5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxd2 | dr11_v1_chr8_+_19674369_19674369 | -0.71 | 1.8e-01 | Click! |
| foxd5 | dr11_v1_chr8_+_30452945_30452945 | -0.70 | 1.9e-01 | Click! |
Activity profile of foxd2_foxd5 motif
Sorted Z-values of foxd2_foxd5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_34127005 | 2.89 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr14_+_21107032 | 2.66 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr4_-_25215968 | 2.37 |
ENSDART00000066932
ENSDART00000066933 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr14_+_11458044 | 2.31 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr18_-_46354269 | 2.15 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr14_+_11457500 | 2.13 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr12_-_22524388 | 2.02 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr21_+_40092301 | 1.90 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr18_-_6803424 | 1.77 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr13_-_4707018 | 1.69 |
ENSDART00000128422
|
oit3
|
oncoprotein induced transcript 3 |
| chr9_-_33063083 | 1.37 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr12_+_46543572 | 1.37 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr7_+_27603211 | 1.37 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr3_-_45848257 | 1.35 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr3_-_45848043 | 1.32 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr21_+_27416284 | 1.27 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr20_-_6532462 | 1.21 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr6_-_55864687 | 1.20 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr22_-_23668356 | 1.18 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr3_-_15081874 | 1.17 |
ENSDART00000192532
|
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr16_+_23975930 | 1.14 |
ENSDART00000147858
ENSDART00000144347 ENSDART00000115270 |
apoc4
|
apolipoprotein C-IV |
| chr5_+_26795465 | 1.14 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr8_-_13013123 | 1.12 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr2_-_40191603 | 1.11 |
ENSDART00000180691
|
si:ch211-122l24.6
|
si:ch211-122l24.6 |
| chr14_-_4120636 | 1.05 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr3_-_50139860 | 1.04 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr14_-_4121052 | 1.04 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr9_-_16109001 | 1.01 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr16_+_23976227 | 0.98 |
ENSDART00000193013
|
apoc4
|
apolipoprotein C-IV |
| chr15_-_37829160 | 0.97 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr18_-_17077596 | 0.96 |
ENSDART00000111821
|
il17c
|
interleukin 17c |
| chr2_+_27855102 | 0.95 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr1_-_30473422 | 0.95 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr18_-_17077419 | 0.94 |
ENSDART00000148714
|
il17c
|
interleukin 17c |
| chr7_+_53156810 | 0.93 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr16_-_13388821 | 0.92 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr6_-_24053404 | 0.91 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr20_-_49889111 | 0.89 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr22_-_38819603 | 0.88 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr20_-_30369598 | 0.87 |
ENSDART00000144549
|
allc
|
allantoicase |
| chr3_+_36424055 | 0.83 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr3_-_29506960 | 0.82 |
ENSDART00000141720
|
cyth4a
|
cytohesin 4a |
| chr16_-_38629208 | 0.81 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr2_-_127945 | 0.81 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr22_-_17595310 | 0.79 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr22_+_11756040 | 0.78 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr7_+_21180747 | 0.77 |
ENSDART00000185543
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr9_-_33081781 | 0.77 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr15_+_45563656 | 0.76 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr12_-_3237561 | 0.75 |
ENSDART00000164665
|
si:ch1073-13h15.3
|
si:ch1073-13h15.3 |
| chr5_-_29238889 | 0.74 |
ENSDART00000143098
|
si:dkey-61l1.4
|
si:dkey-61l1.4 |
| chr9_+_22364997 | 0.74 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr18_-_24988645 | 0.73 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr15_+_45563491 | 0.70 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr22_-_4439311 | 0.69 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr8_+_28695914 | 0.68 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr17_-_10838434 | 0.67 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr3_-_28828242 | 0.66 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr2_-_37956768 | 0.66 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr12_+_2648043 | 0.65 |
ENSDART00000082220
|
gdf2
|
growth differentiation factor 2 |
| chr19_+_9277327 | 0.65 |
ENSDART00000104623
ENSDART00000151164 |
si:rp71-15k1.1
|
si:rp71-15k1.1 |
| chr2_-_37960688 | 0.63 |
ENSDART00000055565
|
cbln14
|
cerebellin 14 |
| chr16_-_19373310 | 0.63 |
ENSDART00000148294
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr19_+_9186175 | 0.63 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr7_+_21841037 | 0.61 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr3_+_15828999 | 0.60 |
ENSDART00000104397
|
tmem11
|
transmembrane protein 11 |
| chr16_-_16619854 | 0.58 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr7_-_44957503 | 0.57 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr16_+_52966812 | 0.57 |
ENSDART00000148435
ENSDART00000049099 ENSDART00000150117 |
trip13
|
thyroid hormone receptor interactor 13 |
| chr6_-_43616936 | 0.55 |
ENSDART00000149301
|
foxp1b
|
forkhead box P1b |
| chr3_+_25191467 | 0.53 |
ENSDART00000156956
ENSDART00000154799 |
il2rb
|
interleukin 2 receptor, beta |
| chr10_+_36650222 | 0.53 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr17_-_27266053 | 0.51 |
ENSDART00000110903
|
E2F2
|
si:ch211-160f23.5 |
| chr1_+_58332000 | 0.50 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr18_-_41161828 | 0.49 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr20_-_22778394 | 0.49 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr22_-_9728208 | 0.48 |
ENSDART00000185962
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr19_+_7043634 | 0.46 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr7_+_29513161 | 0.46 |
ENSDART00000191846
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr18_+_14277003 | 0.45 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr2_+_36898982 | 0.45 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr9_+_3170101 | 0.43 |
ENSDART00000164878
ENSDART00000160567 ENSDART00000161216 |
dync1i2a
|
dynein, cytoplasmic 1, intermediate chain 2a |
| chr2_-_985417 | 0.42 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr8_+_37749263 | 0.42 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr20_-_2641233 | 0.38 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr4_+_57093908 | 0.38 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr8_-_46457233 | 0.38 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr4_-_12723585 | 0.38 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr19_+_32856907 | 0.36 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr21_+_22558187 | 0.35 |
ENSDART00000167599
|
chek1
|
checkpoint kinase 1 |
| chr9_-_12269847 | 0.35 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr5_+_4575800 | 0.34 |
ENSDART00000180952
|
cst14b.2
|
cystatin 14b, tandem duplicate 2 |
| chr7_-_35515931 | 0.33 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr19_+_816208 | 0.33 |
ENSDART00000093304
|
nrm
|
nurim |
| chr10_+_39248911 | 0.33 |
ENSDART00000170079
ENSDART00000167974 |
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr18_+_44532370 | 0.32 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr22_-_13042992 | 0.32 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr25_-_7759453 | 0.31 |
ENSDART00000142439
ENSDART00000021577 |
phf21ab
|
PHD finger protein 21Ab |
| chr24_+_13642126 | 0.31 |
ENSDART00000126769
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr6_+_40437987 | 0.31 |
ENSDART00000136487
|
ghrl
|
ghrelin/obestatin prepropeptide |
| chr22_+_1779401 | 0.30 |
ENSDART00000170126
|
znf1154
|
zinc finger protein 1154 |
| chr21_-_9383974 | 0.30 |
ENSDART00000160932
|
sdad1
|
SDA1 domain containing 1 |
| chr8_-_15150131 | 0.30 |
ENSDART00000138253
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr7_-_17600415 | 0.29 |
ENSDART00000080717
|
nitr6b
|
novel immune-type receptor 6b |
| chr21_-_9384374 | 0.28 |
ENSDART00000169275
|
sdad1
|
SDA1 domain containing 1 |
| chr11_-_42752884 | 0.28 |
ENSDART00000186025
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr7_+_65387374 | 0.28 |
ENSDART00000188680
|
CLEC3A
|
C-type lectin domain family 3 member A |
| chr11_-_18819178 | 0.28 |
ENSDART00000080355
|
zgc:175264
|
zgc:175264 |
| chr6_-_8704702 | 0.27 |
ENSDART00000064149
|
nabp1b
|
nucleic acid binding protein 1b |
| chr5_+_23152282 | 0.27 |
ENSDART00000166783
|
tbx5b
|
T-box 5b |
| chr15_-_16121496 | 0.27 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr19_-_42238440 | 0.26 |
ENSDART00000132591
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr2_+_23007675 | 0.26 |
ENSDART00000163649
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr11_-_20987378 | 0.25 |
ENSDART00000110140
|
taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr15_+_6459847 | 0.25 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr7_-_71585065 | 0.25 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr11_+_14284866 | 0.25 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr7_+_22637515 | 0.24 |
ENSDART00000158698
|
si:dkey-112a7.5
|
si:dkey-112a7.5 |
| chr10_+_26973063 | 0.24 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr20_-_51355465 | 0.24 |
ENSDART00000151620
ENSDART00000151690 ENSDART00000110289 |
tcte1
|
t-complex-associated-testis-expressed 1 |
| chr8_-_16697615 | 0.24 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr4_-_75505048 | 0.24 |
ENSDART00000163665
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr14_+_46410766 | 0.23 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr12_-_30583668 | 0.23 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr15_-_28677725 | 0.23 |
ENSDART00000060255
|
blmh
|
bleomycin hydrolase |
| chr6_-_39919982 | 0.23 |
ENSDART00000065091
ENSDART00000064903 |
sumf1
|
sulfatase modifying factor 1 |
| chr4_-_71913556 | 0.23 |
ENSDART00000167608
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr14_-_28463835 | 0.22 |
ENSDART00000146344
ENSDART00000144076 |
nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr11_-_18818640 | 0.22 |
ENSDART00000191610
ENSDART00000190055 |
zgc:175264
|
zgc:175264 |
| chr14_-_41556720 | 0.22 |
ENSDART00000149244
|
itga6l
|
integrin, alpha 6, like |
| chr9_+_52398531 | 0.22 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr16_-_30564319 | 0.22 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr3_-_12227359 | 0.22 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr16_+_46492994 | 0.22 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr3_-_39208714 | 0.21 |
ENSDART00000125730
|
si:ch211-232p21.6
|
si:ch211-232p21.6 |
| chr20_+_43083745 | 0.21 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr19_+_10536093 | 0.21 |
ENSDART00000138706
|
si:dkey-211g8.1
|
si:dkey-211g8.1 |
| chr14_-_1635296 | 0.21 |
ENSDART00000186658
|
LO018564.1
|
|
| chr5_-_61624693 | 0.21 |
ENSDART00000141323
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr14_-_26377044 | 0.20 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr6_+_2097690 | 0.20 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr18_+_19974289 | 0.19 |
ENSDART00000090334
ENSDART00000192982 |
skor1b
|
SKI family transcriptional corepressor 1b |
| chr3_+_34683096 | 0.18 |
ENSDART00000084432
|
dusp3b
|
dual specificity phosphatase 3b |
| chr2_-_51221198 | 0.18 |
ENSDART00000166869
|
pigrl4.2
|
polymeric immunoglobulin receptor-like 4.2 |
| chr16_+_11724230 | 0.18 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr2_-_6112862 | 0.18 |
ENSDART00000164269
|
prdx1
|
peroxiredoxin 1 |
| chr2_-_51507540 | 0.17 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr2_+_47906240 | 0.17 |
ENSDART00000122206
|
ftr23
|
finTRIM family, member 23 |
| chr3_-_34724879 | 0.17 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr1_-_40140424 | 0.17 |
ENSDART00000148234
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr5_+_54400971 | 0.16 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr5_+_22970617 | 0.16 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr6_-_35051671 | 0.16 |
ENSDART00000132788
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr3_-_16289826 | 0.16 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr21_-_39327223 | 0.16 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr20_+_27647436 | 0.16 |
ENSDART00000180557
ENSDART00000185628 |
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr1_+_532766 | 0.16 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr5_+_33070608 | 0.16 |
ENSDART00000135063
|
npffr2b
|
neuropeptide FF receptor 2b |
| chr20_+_26881600 | 0.15 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr19_-_42238003 | 0.15 |
ENSDART00000151022
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr6_+_41186320 | 0.15 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr11_+_8660158 | 0.15 |
ENSDART00000169141
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr10_+_26597990 | 0.15 |
ENSDART00000079187
|
fhl1b
|
four and a half LIM domains 1b |
| chr8_+_29636431 | 0.15 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr12_+_23424108 | 0.15 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr16_-_1503023 | 0.14 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr8_+_23142946 | 0.14 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr20_-_52603094 | 0.14 |
ENSDART00000136945
ENSDART00000133798 |
si:dkey-235d18.6
|
si:dkey-235d18.6 |
| chr1_-_21287724 | 0.13 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr5_+_35398745 | 0.13 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr4_-_6373735 | 0.13 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr22_+_31059919 | 0.13 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr6_-_8264751 | 0.13 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr16_-_7692568 | 0.12 |
ENSDART00000148980
|
pimr209
|
Pim proto-oncogene, serine/threonine kinase, related 209 |
| chr22_-_5663354 | 0.12 |
ENSDART00000081774
|
ccdc51
|
coiled-coil domain containing 51 |
| chr6_-_35052145 | 0.12 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr5_-_29671586 | 0.12 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr23_+_28381260 | 0.12 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr7_+_67429185 | 0.11 |
ENSDART00000162553
ENSDART00000178646 |
kars
|
lysyl-tRNA synthetase |
| chr19_+_11984725 | 0.11 |
ENSDART00000185960
|
spag1a
|
sperm associated antigen 1a |
| chr17_-_10073926 | 0.11 |
ENSDART00000166081
ENSDART00000161574 |
zgc:109986
|
zgc:109986 |
| chr12_-_20120702 | 0.11 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr8_+_19624589 | 0.10 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr17_+_26828027 | 0.10 |
ENSDART00000042060
|
jade1
|
jade family PHD finger 1 |
| chr2_-_5135125 | 0.10 |
ENSDART00000164039
|
ptmab
|
prothymosin, alpha b |
| chr14_-_33613794 | 0.10 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr8_-_47329755 | 0.10 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr5_-_51756210 | 0.10 |
ENSDART00000163464
|
lhfpl2b
|
LHFPL tetraspan subfamily member 2b |
| chr20_+_39375280 | 0.10 |
ENSDART00000172434
|
pimr131
|
Pim proto-oncogene, serine/threonine kinase, related 131 |
| chr4_-_40883162 | 0.10 |
ENSDART00000151978
|
znf1102
|
zinc finger protein 1102 |
| chr15_+_17623115 | 0.09 |
ENSDART00000062378
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr4_-_45337903 | 0.09 |
ENSDART00000159767
|
si:dkey-247i3.6
|
si:dkey-247i3.6 |
| chr5_-_67750907 | 0.09 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr17_+_30448452 | 0.09 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr10_+_26972755 | 0.09 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr15_+_25156382 | 0.09 |
ENSDART00000171092
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr7_-_58776400 | 0.09 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr16_+_43139504 | 0.08 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr10_-_22491353 | 0.08 |
ENSDART00000180783
ENSDART00000159564 |
epob
|
erythropoietin b |
| chr24_+_7631797 | 0.08 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd2_foxd5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.4 | 1.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.1 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.3 | 0.9 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.2 | 1.4 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.2 | 1.2 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 1.0 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.3 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 1.0 | GO:0044206 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 1.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.3 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.2 | GO:0050817 | coagulation(GO:0050817) regulation of coagulation(GO:0050818) negative regulation of coagulation(GO:0050819) |
| 0.1 | 1.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.2 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.7 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.7 | GO:0032095 | regulation of response to food(GO:0032095) |
| 0.1 | 0.5 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.2 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 1.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.5 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.0 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 0.6 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.9 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 3.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 3.0 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 0.2 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0006999 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.3 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 3.0 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.3 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 0.8 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 1.0 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.3 | 2.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.7 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.2 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.5 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 1.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 1.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 1.0 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 2.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 2.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 1.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.3 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 2.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |