Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
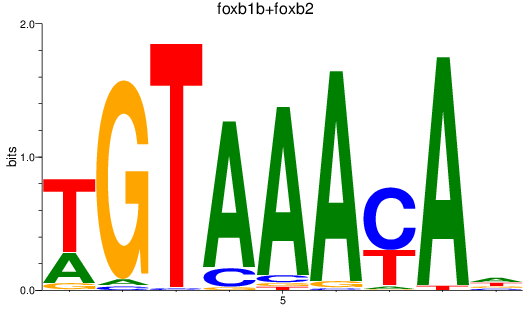
Results for foxb1b+foxb2
Z-value: 1.51
Transcription factors associated with foxb1b+foxb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxb2
|
ENSDARG00000037475 | forkhead box B2 |
|
foxb1b
|
ENSDARG00000053650 | forkhead box B1b |
|
foxb1b
|
ENSDARG00000110408 | forkhead box B1b |
|
foxb1b
|
ENSDARG00000113373 | forkhead box B1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxb2 | dr11_v1_chr8_-_38506339_38506339 | 0.84 | 7.6e-02 | Click! |
| foxb1b | dr11_v1_chr7_+_29461060_29461060 | 0.45 | 4.4e-01 | Click! |
Activity profile of foxb1b+foxb2 motif
Sorted Z-values of foxb1b+foxb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_19953089 | 1.57 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr2_-_8017579 | 1.28 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr6_+_40671336 | 1.20 |
ENSDART00000111639
ENSDART00000186617 |
rereb
|
arginine-glutamic acid dipeptide (RE) repeats b |
| chr12_+_28367557 | 1.18 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr21_+_11684830 | 1.08 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr23_-_29502287 | 1.03 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr21_+_11685009 | 1.02 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr7_+_22823889 | 1.01 |
ENSDART00000127467
ENSDART00000148576 ENSDART00000149993 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr1_+_16127825 | 0.96 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr23_-_3703569 | 0.95 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr1_+_25801648 | 0.94 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr20_-_34801181 | 0.93 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr7_+_48288762 | 0.91 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr6_-_9792004 | 0.90 |
ENSDART00000081129
|
cdk15
|
cyclin-dependent kinase 15 |
| chr23_-_27633730 | 0.89 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr17_-_20287530 | 0.88 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr14_-_7885707 | 0.86 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr15_+_8043751 | 0.83 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr4_+_8797197 | 0.82 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr2_+_26240339 | 0.82 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr3_+_34821327 | 0.80 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr8_+_41533268 | 0.79 |
ENSDART00000142377
|
si:ch211-158d24.2
|
si:ch211-158d24.2 |
| chr15_-_18574716 | 0.79 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr11_+_6819050 | 0.79 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr5_+_52625975 | 0.78 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr2_-_24462277 | 0.78 |
ENSDART00000033922
|
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr2_+_26240631 | 0.78 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr6_-_40744720 | 0.76 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr11_-_29833698 | 0.76 |
ENSDART00000079149
|
xk
|
X-linked Kx blood group (McLeod syndrome) |
| chr5_-_40190949 | 0.75 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr2_+_24177006 | 0.75 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr19_-_28367413 | 0.75 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr6_+_36942966 | 0.75 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr10_+_34426256 | 0.75 |
ENSDART00000102566
|
nbeaa
|
neurobeachin a |
| chr21_-_29100110 | 0.74 |
ENSDART00000142598
|
timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr14_+_17197132 | 0.74 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr16_-_12173399 | 0.73 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr16_-_12173554 | 0.71 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr3_+_19207176 | 0.71 |
ENSDART00000087803
|
rln3a
|
relaxin 3a |
| chr3_-_60142530 | 0.71 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr16_-_43025885 | 0.70 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr9_+_38962398 | 0.69 |
ENSDART00000134294
|
map2
|
microtubule-associated protein 2 |
| chr19_+_6938289 | 0.69 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr21_-_30284404 | 0.69 |
ENSDART00000066363
|
zgc:175066
|
zgc:175066 |
| chr4_-_9586713 | 0.68 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr6_-_13187168 | 0.67 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr21_+_30563115 | 0.66 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr8_-_25120231 | 0.65 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr2_-_40135942 | 0.65 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr14_+_26796684 | 0.64 |
ENSDART00000187414
|
klhl4
|
kelch-like family member 4 |
| chr14_-_4682114 | 0.64 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr5_-_46896541 | 0.64 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr18_+_17428506 | 0.62 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr18_-_39787040 | 0.62 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr19_+_10396042 | 0.62 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr9_+_3388099 | 0.61 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr12_+_18578597 | 0.60 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr10_-_15405564 | 0.59 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr6_-_32999646 | 0.59 |
ENSDART00000159510
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr13_-_21701323 | 0.59 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr24_-_4973765 | 0.59 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr23_-_29505645 | 0.59 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr3_-_28428198 | 0.58 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr25_+_19149241 | 0.58 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr23_-_15330168 | 0.57 |
ENSDART00000035865
ENSDART00000143635 |
sulf2b
|
sulfatase 2b |
| chr3_-_51912019 | 0.57 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr17_-_40397752 | 0.57 |
ENSDART00000178483
|
BX548062.1
|
|
| chr12_+_31729075 | 0.57 |
ENSDART00000152973
|
RNF157
|
si:dkey-49c17.3 |
| chr17_-_3986236 | 0.57 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr12_-_4781801 | 0.57 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr7_-_31941670 | 0.57 |
ENSDART00000180929
ENSDART00000075389 |
bdnf
|
brain-derived neurotrophic factor |
| chr1_+_53954230 | 0.57 |
ENSDART00000037729
ENSDART00000159900 |
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr4_+_6643421 | 0.56 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr11_-_4235811 | 0.56 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr19_+_16032383 | 0.56 |
ENSDART00000046530
|
rab42a
|
RAB42, member RAS oncogene family a |
| chr8_-_9118958 | 0.56 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr8_-_18582922 | 0.56 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr18_+_19419120 | 0.55 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr22_+_27090136 | 0.55 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr23_+_19594608 | 0.55 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr20_-_19590378 | 0.55 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr4_+_8168514 | 0.54 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr6_+_13933464 | 0.54 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr10_+_34426571 | 0.54 |
ENSDART00000144529
|
nbeaa
|
neurobeachin a |
| chr25_+_10458990 | 0.54 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr18_-_14734678 | 0.53 |
ENSDART00000142462
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr17_-_37474689 | 0.52 |
ENSDART00000103980
|
crip2
|
cysteine-rich protein 2 |
| chr2_+_24177190 | 0.52 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr23_+_16620801 | 0.52 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr22_+_20195280 | 0.52 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr13_+_1100197 | 0.52 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr14_-_49063157 | 0.52 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr23_-_29505463 | 0.51 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr7_+_48460239 | 0.51 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr13_-_32995324 | 0.51 |
ENSDART00000140542
ENSDART00000037740 |
kcnf1b
|
potassium voltage-gated channel, subfamily F, member 1b |
| chr6_+_23752593 | 0.51 |
ENSDART00000164366
|
zgc:158654
|
zgc:158654 |
| chr14_-_26177156 | 0.50 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr25_+_3677650 | 0.50 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr14_-_34044369 | 0.50 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_+_5604241 | 0.50 |
ENSDART00000011025
|
wipf2b
|
WAS/WASL interacting protein family, member 2b |
| chr15_+_37105986 | 0.50 |
ENSDART00000157762
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr3_-_28258462 | 0.50 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr1_-_40994259 | 0.50 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr3_-_26341959 | 0.50 |
ENSDART00000169344
ENSDART00000142878 ENSDART00000087196 |
zgc:153240
|
zgc:153240 |
| chr20_-_26039841 | 0.49 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr8_-_32497815 | 0.49 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr6_-_39605734 | 0.49 |
ENSDART00000044276
ENSDART00000179059 |
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr21_+_44684577 | 0.49 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr16_-_17162843 | 0.49 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr20_-_19365875 | 0.49 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr25_+_15647993 | 0.48 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr20_+_30445971 | 0.48 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr5_-_46980651 | 0.48 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr2_+_38924975 | 0.48 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr20_+_40457599 | 0.48 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr18_-_8579907 | 0.48 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr15_-_22147860 | 0.47 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr9_-_46072805 | 0.47 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr16_-_52879741 | 0.47 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr6_+_13787855 | 0.47 |
ENSDART00000182899
|
tmem198b
|
transmembrane protein 198b |
| chr16_+_30117798 | 0.47 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr7_+_36898850 | 0.47 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr13_+_23132666 | 0.46 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr21_-_4032650 | 0.46 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr4_+_12113105 | 0.46 |
ENSDART00000182399
|
tmem178b
|
transmembrane protein 178B |
| chr7_+_7019911 | 0.46 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr19_+_24039830 | 0.46 |
ENSDART00000100422
|
rit1
|
Ras-like without CAAX 1 |
| chr1_-_10473630 | 0.46 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr8_-_32497581 | 0.46 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr23_+_31405497 | 0.46 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr6_-_30658755 | 0.45 |
ENSDART00000065215
ENSDART00000181302 |
lurap1
|
leucine rich adaptor protein 1 |
| chr6_+_12006557 | 0.45 |
ENSDART00000128024
|
wdsub1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr19_-_7358184 | 0.45 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr12_+_31729498 | 0.45 |
ENSDART00000188546
ENSDART00000182562 ENSDART00000186147 |
RNF157
|
si:dkey-49c17.3 |
| chr18_+_24922125 | 0.45 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr10_+_25219728 | 0.45 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr16_+_46000956 | 0.45 |
ENSDART00000101753
ENSDART00000162393 |
mtmr11
|
myotubularin related protein 11 |
| chr4_+_17280868 | 0.45 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr2_+_25929619 | 0.44 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr2_-_11662851 | 0.44 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr13_-_40499296 | 0.44 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr11_+_30162407 | 0.44 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr13_+_1944451 | 0.43 |
ENSDART00000125914
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr9_-_44295071 | 0.43 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr5_-_68495224 | 0.42 |
ENSDART00000187955
|
ephb4a
|
eph receptor B4a |
| chr5_+_52705539 | 0.41 |
ENSDART00000189436
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr9_+_29603649 | 0.41 |
ENSDART00000140477
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr2_-_30668580 | 0.41 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr17_+_31221761 | 0.41 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr5_-_45634675 | 0.41 |
ENSDART00000168534
|
NPFFR2
|
neuropeptide FF receptor 2a |
| chr11_-_36341189 | 0.41 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr13_+_32446169 | 0.41 |
ENSDART00000143325
|
nt5c1ba
|
5'-nucleotidase, cytosolic IB a |
| chr2_+_20472150 | 0.41 |
ENSDART00000168537
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr12_+_27704015 | 0.41 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr12_+_32729470 | 0.40 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr12_+_22404108 | 0.40 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr6_-_957830 | 0.40 |
ENSDART00000090019
ENSDART00000184286 |
zeb2b
|
zinc finger E-box binding homeobox 2b |
| chr3_-_6759534 | 0.40 |
ENSDART00000180862
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr17_-_21278846 | 0.40 |
ENSDART00000181356
|
hspa12a
|
heat shock protein 12A |
| chr13_-_18691041 | 0.40 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr1_+_6171585 | 0.40 |
ENSDART00000024358
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr18_+_7286788 | 0.40 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr9_+_38163876 | 0.40 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr15_-_18138607 | 0.39 |
ENSDART00000176690
|
CR385077.1
|
|
| chr17_+_25290136 | 0.39 |
ENSDART00000173295
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr5_-_38384289 | 0.39 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr1_+_2129164 | 0.39 |
ENSDART00000074923
ENSDART00000124534 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr13_-_49169545 | 0.39 |
ENSDART00000192076
|
tsnax
|
translin-associated factor X |
| chr14_+_29581710 | 0.39 |
ENSDART00000188820
ENSDART00000193874 |
TENM2
|
si:dkey-34l15.2 |
| chr2_-_50372467 | 0.39 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr9_+_33154841 | 0.39 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr6_-_13408680 | 0.39 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr24_-_21989406 | 0.39 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr14_-_2352384 | 0.38 |
ENSDART00000170666
|
si:ch73-233f7.7
|
si:ch73-233f7.7 |
| chr18_-_13121983 | 0.38 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr16_-_7828838 | 0.38 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr1_+_2128970 | 0.38 |
ENSDART00000180074
ENSDART00000022019 ENSDART00000098059 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr8_+_36500061 | 0.38 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr5_-_37875636 | 0.38 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr22_-_25033105 | 0.38 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr9_-_3149896 | 0.38 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr9_+_19623363 | 0.38 |
ENSDART00000142471
ENSDART00000147662 ENSDART00000136053 |
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr13_-_11378355 | 0.37 |
ENSDART00000164566
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr24_-_22756508 | 0.37 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr25_+_388258 | 0.37 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr7_+_20110336 | 0.37 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr16_-_26437668 | 0.37 |
ENSDART00000142056
|
megf8
|
multiple EGF-like-domains 8 |
| chr5_+_34981584 | 0.37 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr1_-_10048514 | 0.37 |
ENSDART00000125358
ENSDART00000054835 |
rnf175
|
ring finger protein 175 |
| chr19_-_27261102 | 0.37 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr18_-_5781922 | 0.36 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr11_+_7580079 | 0.36 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr14_-_17599452 | 0.36 |
ENSDART00000080042
|
rab33a
|
RAB33A, member RAS oncogene family |
| chr17_+_39790388 | 0.36 |
ENSDART00000149488
|
ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr20_+_42918755 | 0.36 |
ENSDART00000134855
|
efr3bb
|
EFR3 homolog Bb (S. cerevisiae) |
| chr4_-_16412084 | 0.36 |
ENSDART00000188460
|
dcn
|
decorin |
| chr24_+_19210001 | 0.36 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr1_-_8917902 | 0.36 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr9_+_2343096 | 0.36 |
ENSDART00000062292
ENSDART00000191722 ENSDART00000135180 |
atf2
|
activating transcription factor 2 |
| chr15_+_35718354 | 0.36 |
ENSDART00000110003
|
nyap2a
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxb1b+foxb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.3 | 3.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 1.0 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.3 | 1.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 0.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 0.9 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.2 | 0.7 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.2 | 0.6 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 0.6 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.4 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 2.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.5 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.1 | 0.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.5 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 0.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.5 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.6 | GO:0048853 | cell proliferation in hindbrain(GO:0021534) forebrain morphogenesis(GO:0048853) |
| 0.1 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.6 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.2 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.4 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.4 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.1 | 0.2 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.3 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.3 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.4 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.9 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.5 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.7 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.9 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.7 | GO:0097502 | protein mannosylation(GO:0035268) mannosylation(GO:0097502) |
| 0.0 | 0.2 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.4 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 1.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.2 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.9 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.4 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 1.7 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.0 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.0 | 0.6 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.3 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.0 | 0.1 | GO:0045830 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 1.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.9 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 1.3 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.4 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 0.6 | GO:0072380 | TRC complex(GO:0072380) |
| 0.2 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.7 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.9 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 2.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 3.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 2.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.3 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 6.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.0 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 0.5 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.2 | 2.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.9 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.4 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.3 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.4 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.3 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.7 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) inositol trisphosphate kinase activity(GO:0051766) |
| 0.1 | 0.2 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 1.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 4.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.2 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |