Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
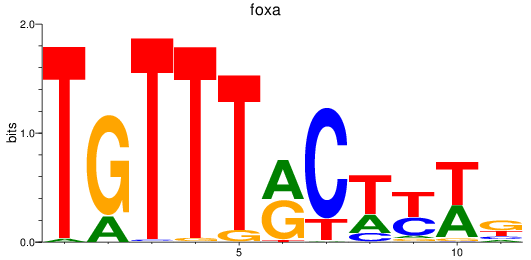
Results for foxa_foxa1_foxa2
Z-value: 2.38
Transcription factors associated with foxa_foxa1_foxa2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxa
|
ENSDARG00000087094 | forkhead box A sequence |
|
foxa
|
ENSDARG00000110743 | forkhead box A sequence |
|
foxa
|
ENSDARG00000115019 | forkhead box A sequence |
|
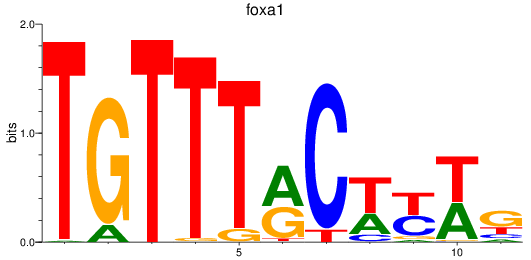
foxa1
|
ENSDARG00000102138 | forkhead box A1 |
|
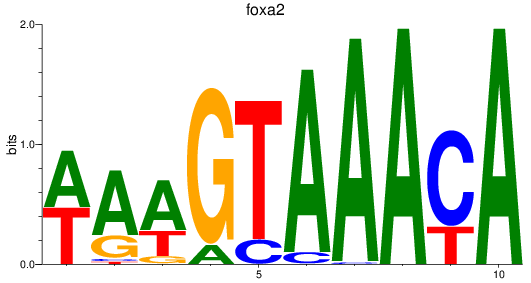
foxa2
|
ENSDARG00000003411 | forkhead box A2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa2 | dr11_v1_chr17_-_42568498_42568498 | 0.82 | 9.0e-02 | Click! |
| foxa | dr11_v1_chr14_-_33981544_33981544 | 0.34 | 5.7e-01 | Click! |
| foxa1 | dr11_v1_chr17_+_10318071_10318071 | 0.31 | 6.1e-01 | Click! |
Activity profile of foxa_foxa1_foxa2 motif
Sorted Z-values of foxa_foxa1_foxa2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24967348 | 3.12 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr12_+_31616412 | 3.11 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr20_+_6142433 | 2.97 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr6_-_55864687 | 2.51 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr16_-_21785261 | 2.50 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr16_-_54455573 | 2.38 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr22_-_36856405 | 2.33 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr23_+_28770225 | 2.28 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr2_+_10134345 | 2.23 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr5_+_28857969 | 2.13 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr5_+_28797771 | 2.11 |
ENSDART00000188845
ENSDART00000149066 |
si:ch211-186e20.7
|
si:ch211-186e20.7 |
| chr10_+_26747755 | 1.86 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr16_+_23978978 | 1.82 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr10_+_17026870 | 1.80 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr5_+_28830643 | 1.77 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_+_28830388 | 1.68 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_+_28848870 | 1.67 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr4_-_16824231 | 1.67 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr5_+_28849155 | 1.65 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr4_-_16824556 | 1.60 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr15_+_14856307 | 1.51 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr7_+_69019851 | 1.50 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr5_+_28858345 | 1.37 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr14_+_15155684 | 1.32 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr20_+_23440632 | 1.28 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr2_+_16781015 | 1.25 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr2_+_16780643 | 1.21 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr16_-_31661536 | 1.17 |
ENSDART00000169973
|
wu:fd46c06
|
wu:fd46c06 |
| chr23_+_44881020 | 1.15 |
ENSDART00000149355
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr10_+_7709724 | 1.12 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr17_+_24036791 | 1.09 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr4_+_18806251 | 1.05 |
ENSDART00000138662
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr4_+_1530287 | 1.04 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr23_-_25686894 | 1.03 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr15_-_25365570 | 1.01 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr24_+_22056386 | 0.93 |
ENSDART00000132390
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr24_+_26887487 | 0.90 |
ENSDART00000189425
|
CR376848.1
|
|
| chr13_+_23988442 | 0.90 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr4_+_5868034 | 0.88 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr6_+_6780873 | 0.88 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr12_-_22238004 | 0.88 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr4_+_17310143 | 0.86 |
ENSDART00000004717
|
igf1
|
insulin-like growth factor 1 |
| chr21_+_30502002 | 0.83 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr5_-_66160415 | 0.82 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr18_-_40901707 | 0.80 |
ENSDART00000139352
|
foxg1c
|
forkhead box G1c |
| chr15_+_9121054 | 0.79 |
ENSDART00000193889
|
ptgir
|
prostaglandin I2 receptor |
| chr5_-_4931266 | 0.76 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr11_-_4235811 | 0.76 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr6_-_9565526 | 0.74 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr8_-_40075983 | 0.72 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr2_-_24068848 | 0.72 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr8_+_49570884 | 0.71 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr8_+_30709685 | 0.69 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr1_-_9134045 | 0.69 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr16_+_14588141 | 0.67 |
ENSDART00000140469
ENSDART00000059984 ENSDART00000167411 ENSDART00000133566 |
deptor
|
DEP domain containing MTOR-interacting protein |
| chr21_+_27416284 | 0.67 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr2_+_1486822 | 0.67 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr20_-_36809059 | 0.67 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr2_+_1487118 | 0.67 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr3_+_19299309 | 0.66 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr2_+_11028923 | 0.66 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr8_-_29822527 | 0.65 |
ENSDART00000167487
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr23_-_31763753 | 0.65 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr15_-_30816370 | 0.62 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr24_+_7782313 | 0.62 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr18_-_24988645 | 0.61 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr13_-_31647323 | 0.61 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr19_+_30990815 | 0.61 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr5_+_65991152 | 0.61 |
ENSDART00000097756
|
lcn15
|
lipocalin 15 |
| chr15_-_25365319 | 0.60 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr11_-_3629201 | 0.58 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr21_+_40092301 | 0.57 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr22_-_14128716 | 0.57 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr12_-_25294096 | 0.57 |
ENSDART00000183398
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr25_-_12805295 | 0.56 |
ENSDART00000157629
|
ca5a
|
carbonic anhydrase Va |
| chr7_-_71384391 | 0.56 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr2_-_38035235 | 0.56 |
ENSDART00000075904
|
cbln5
|
cerebellin 5 |
| chr8_-_35960987 | 0.56 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr18_-_6803424 | 0.55 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr14_+_32918484 | 0.55 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr20_+_6756247 | 0.55 |
ENSDART00000167344
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr24_+_20575259 | 0.55 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr7_+_67699178 | 0.54 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr2_+_11029138 | 0.54 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr17_+_16755287 | 0.53 |
ENSDART00000080129
|
ston2
|
stonin 2 |
| chr24_-_26369185 | 0.53 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr17_-_26867725 | 0.52 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr9_-_19161982 | 0.51 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr13_-_40726865 | 0.51 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr8_-_25771474 | 0.49 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr7_+_9904627 | 0.49 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr7_-_31759394 | 0.49 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr21_-_26028205 | 0.48 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr16_-_19373310 | 0.48 |
ENSDART00000148294
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr21_-_39081107 | 0.48 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr15_-_18240199 | 0.48 |
ENSDART00000109999
|
or131-2
|
odorant receptor, family H, subfamily 131, member 2 |
| chr23_+_21966447 | 0.47 |
ENSDART00000189378
|
lactbl1a
|
lactamase, beta-like 1a |
| chr9_+_52411530 | 0.47 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr5_+_36513605 | 0.47 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr17_-_15188440 | 0.46 |
ENSDART00000151885
|
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr23_+_24085531 | 0.46 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr4_-_6373735 | 0.45 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr18_-_44847855 | 0.45 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr5_+_32345187 | 0.44 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr9_+_11293830 | 0.44 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr10_-_3416258 | 0.44 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr23_-_33750307 | 0.43 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr10_+_41199660 | 0.43 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr19_-_46091497 | 0.42 |
ENSDART00000178772
ENSDART00000167255 |
ptdss1b
si:dkey-108k24.2
|
phosphatidylserine synthase 1b si:dkey-108k24.2 |
| chr22_-_26236188 | 0.42 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr15_+_36187434 | 0.42 |
ENSDART00000181536
ENSDART00000099501 ENSDART00000154432 |
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr15_-_4011013 | 0.42 |
ENSDART00000158029
|
CR626884.3
|
|
| chr19_-_7272921 | 0.42 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr16_-_44673851 | 0.42 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr17_-_30652738 | 0.42 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr5_+_45677781 | 0.40 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr12_-_28537615 | 0.40 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr25_-_35524166 | 0.40 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr8_+_22516728 | 0.39 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr23_-_33750135 | 0.39 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr20_-_23440955 | 0.39 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr11_+_13223625 | 0.39 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr16_-_30847192 | 0.38 |
ENSDART00000191106
ENSDART00000128158 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr19_+_7636941 | 0.38 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr9_+_53725128 | 0.38 |
ENSDART00000169062
|
cnmd
|
chondromodulin |
| chr2_-_43168292 | 0.37 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr15_-_18232712 | 0.37 |
ENSDART00000081199
|
wu:fj20b03
|
wu:fj20b03 |
| chr1_+_51537250 | 0.37 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr9_+_50110763 | 0.37 |
ENSDART00000162990
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr5_-_64103863 | 0.36 |
ENSDART00000135014
ENSDART00000083684 |
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr16_+_52105227 | 0.36 |
ENSDART00000150025
ENSDART00000097863 |
MAN1C1
|
si:ch73-373m9.1 |
| chr5_-_29238889 | 0.36 |
ENSDART00000143098
|
si:dkey-61l1.4
|
si:dkey-61l1.4 |
| chr15_+_38299385 | 0.36 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr15_+_38299563 | 0.36 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr17_-_52587598 | 0.35 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr13_-_33207367 | 0.35 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr22_-_6066867 | 0.35 |
ENSDART00000142383
|
si:dkey-19a16.1
|
si:dkey-19a16.1 |
| chr9_-_38036984 | 0.35 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr2_-_5475910 | 0.35 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr16_+_19029297 | 0.35 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr16_-_17713859 | 0.34 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr11_+_35171406 | 0.34 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr16_-_25606235 | 0.34 |
ENSDART00000192741
|
zgc:110410
|
zgc:110410 |
| chr1_-_25177086 | 0.34 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr13_-_16066997 | 0.34 |
ENSDART00000184790
|
SPATA48
|
spermatogenesis associated 48 |
| chr9_+_53725294 | 0.34 |
ENSDART00000165991
|
cnmd
|
chondromodulin |
| chr11_-_19576890 | 0.33 |
ENSDART00000157847
|
atxn7
|
ataxin 7 |
| chr8_+_17184602 | 0.32 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr16_-_34477805 | 0.32 |
ENSDART00000136546
|
serinc2l
|
serine incorporator 2, like |
| chr4_-_17725008 | 0.32 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr20_+_52546186 | 0.32 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr3_-_55525627 | 0.32 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr20_+_27003558 | 0.32 |
ENSDART00000125688
|
si:dkey-177p2.18
|
si:dkey-177p2.18 |
| chr2_+_23731194 | 0.32 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr24_-_31306724 | 0.31 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr6_+_40775800 | 0.31 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr4_+_14981854 | 0.31 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr14_+_8710568 | 0.31 |
ENSDART00000169505
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr6_+_27418541 | 0.30 |
ENSDART00000187410
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr20_-_10288156 | 0.30 |
ENSDART00000064110
|
si:dkey-63b1.1
|
si:dkey-63b1.1 |
| chr8_-_25605537 | 0.30 |
ENSDART00000005906
|
stk38a
|
serine/threonine kinase 38a |
| chr15_+_20843161 | 0.30 |
ENSDART00000153726
|
ulk2
|
unc-51 like autophagy activating kinase 2 |
| chr7_-_66137065 | 0.30 |
ENSDART00000145843
|
pth1b
|
parathyroid hormone 1b |
| chr16_-_25606889 | 0.30 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr15_+_17730567 | 0.30 |
ENSDART00000180790
|
si:ch211-213d14.1
|
si:ch211-213d14.1 |
| chr12_-_1361517 | 0.29 |
ENSDART00000188297
|
LO018020.2
|
|
| chr2_-_5466708 | 0.29 |
ENSDART00000136682
|
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr14_-_6931889 | 0.29 |
ENSDART00000166439
|
si:ch211-266k2.1
|
si:ch211-266k2.1 |
| chr18_+_17611627 | 0.29 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr14_-_36799280 | 0.29 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr3_+_46762703 | 0.29 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr9_-_6991650 | 0.28 |
ENSDART00000081718
|
slc9a2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr21_+_10866421 | 0.28 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr20_-_20533865 | 0.28 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr11_-_7411436 | 0.28 |
ENSDART00000167312
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr11_-_35171162 | 0.27 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr22_+_105376 | 0.27 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr8_-_13029297 | 0.27 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr16_-_30564319 | 0.27 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr16_+_28881235 | 0.27 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr5_-_33236637 | 0.27 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr14_-_30390145 | 0.27 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr13_+_18545819 | 0.27 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr15_-_31372486 | 0.27 |
ENSDART00000127447
|
or111-4
|
odorant receptor, family D, subfamily 111, member 4 |
| chr19_-_24958393 | 0.26 |
ENSDART00000098592
|
si:ch211-195b13.6
|
si:ch211-195b13.6 |
| chr22_-_3914162 | 0.26 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
| chr1_-_31856622 | 0.26 |
ENSDART00000065125
|
nt5c2b
|
5'-nucleotidase, cytosolic IIb |
| chr10_-_5135788 | 0.26 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr4_-_75157223 | 0.26 |
ENSDART00000174127
|
CABZ01066312.1
|
|
| chr18_-_7448047 | 0.26 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr14_+_32926385 | 0.26 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr9_-_41062412 | 0.26 |
ENSDART00000193879
|
ankar
|
ankyrin and armadillo repeat containing |
| chr4_-_19921410 | 0.26 |
ENSDART00000184359
ENSDART00000179965 |
BX323820.1
|
|
| chr13_-_8692860 | 0.26 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr22_+_17784414 | 0.26 |
ENSDART00000188189
ENSDART00000145260 |
tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr8_-_16725959 | 0.26 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr6_-_30683637 | 0.26 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr7_+_21180747 | 0.26 |
ENSDART00000185543
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr5_-_34875858 | 0.25 |
ENSDART00000085086
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr11_+_18612421 | 0.25 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa_foxa1_foxa2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.8 | 3.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 2.4 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.3 | 1.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.3 | 3.0 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.2 | 2.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 0.9 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.6 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.2 | 1.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 1.1 | GO:0043091 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 14.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 3.5 | GO:0009749 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 2.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.8 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 1.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.8 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.7 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.4 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.3 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.7 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 3.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.3 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0006971 | hypotonic response(GO:0006971) hypotonic salinity response(GO:0042539) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 2.5 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.1 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 2.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.5 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.3 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.2 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.0 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 1.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.8 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.0 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 0.5 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.4 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 3.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 23.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.7 | 3.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.6 | 2.5 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.4 | 2.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 0.7 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 1.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 13.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.4 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.3 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 3.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.7 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.3 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.8 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.1 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.3 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0050997 | phosphatidylcholine binding(GO:0031210) quaternary ammonium group binding(GO:0050997) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 2.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 7.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 1.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 0.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 2.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 3.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |