Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
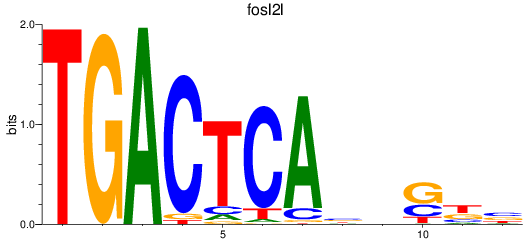
Results for fosl2l
Z-value: 0.56
Transcription factors associated with fosl2l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2l
|
ENSDARG00000092174 | fos-like antigen 2 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-59d15.4 | dr11_v1_chr20_+_9211237_9211237 | -0.44 | 4.6e-01 | Click! |
Activity profile of fosl2l motif
Sorted Z-values of fosl2l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23403602 | 0.59 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr7_-_7398350 | 0.47 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr18_+_47313715 | 0.40 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr18_+_47313899 | 0.38 |
ENSDART00000192389
ENSDART00000189592 ENSDART00000184281 |
barx2
|
BARX homeobox 2 |
| chr23_-_1017605 | 0.38 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr2_+_38731696 | 0.37 |
ENSDART00000181733
|
FQ377660.1
|
|
| chr4_+_77661056 | 0.37 |
ENSDART00000152953
|
si:dkey-61p9.7
|
si:dkey-61p9.7 |
| chr13_-_33134611 | 0.36 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr25_-_13408760 | 0.36 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr6_+_54711306 | 0.35 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr19_-_7420867 | 0.35 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr10_+_38593645 | 0.35 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr23_-_1017428 | 0.35 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr7_-_8417315 | 0.34 |
ENSDART00000173046
|
jac1
|
jacalin 1 |
| chr25_+_10410620 | 0.33 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr5_+_9037650 | 0.33 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr8_+_8671229 | 0.32 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr3_+_21189766 | 0.30 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr8_-_38201415 | 0.28 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr20_-_35578435 | 0.27 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr7_+_39706004 | 0.26 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr24_+_38223618 | 0.26 |
ENSDART00000003495
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr3_-_58831683 | 0.26 |
ENSDART00000110292
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr16_+_49005321 | 0.25 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr20_+_28364742 | 0.22 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr2_-_985417 | 0.22 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr8_+_6576940 | 0.22 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr16_+_26547152 | 0.21 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr5_-_1047504 | 0.21 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr1_-_25177086 | 0.20 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr3_+_34121156 | 0.20 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr9_+_2499627 | 0.20 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr3_-_48259289 | 0.19 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr22_+_19247255 | 0.19 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr20_+_53522059 | 0.19 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr20_+_26880668 | 0.19 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr6_-_609880 | 0.19 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr25_-_33275666 | 0.19 |
ENSDART00000193605
|
CABZ01046954.1
|
|
| chr17_+_51744450 | 0.19 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr20_-_34127415 | 0.19 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr25_+_13205878 | 0.18 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr10_-_44482911 | 0.18 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr20_-_26531850 | 0.18 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr7_+_19482084 | 0.18 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr9_-_2594410 | 0.18 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr8_-_18667693 | 0.18 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr17_+_6585333 | 0.17 |
ENSDART00000124293
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr1_+_57041549 | 0.17 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr15_-_33834577 | 0.17 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr1_-_52498146 | 0.17 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr24_+_30215475 | 0.17 |
ENSDART00000164717
|
si:ch73-358j7.2
|
si:ch73-358j7.2 |
| chr23_-_45398622 | 0.17 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr20_-_26532167 | 0.17 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr21_-_27338639 | 0.17 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr24_+_38216340 | 0.16 |
ENSDART00000188561
|
CU896602.4
|
|
| chr23_+_28378543 | 0.16 |
ENSDART00000145327
|
zgc:153867
|
zgc:153867 |
| chr17_+_51743908 | 0.16 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr20_-_34670236 | 0.16 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr25_+_5972690 | 0.16 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr8_-_11170114 | 0.16 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr13_-_15982707 | 0.16 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr15_+_42235449 | 0.16 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr1_-_56176976 | 0.16 |
ENSDART00000052688
|
c3a.1
|
complement component c3a, duplicate 1 |
| chr13_-_20381485 | 0.16 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr3_+_4346854 | 0.15 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr19_+_48359259 | 0.15 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr16_+_41517188 | 0.15 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr22_+_38194151 | 0.15 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr10_+_38610741 | 0.15 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr14_-_22100118 | 0.15 |
ENSDART00000157547
|
ssrp1a
|
structure specific recognition protein 1a |
| chr4_+_16715267 | 0.15 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr15_-_18197008 | 0.15 |
ENSDART00000147632
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr5_+_22370167 | 0.15 |
ENSDART00000145163
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr23_-_36316352 | 0.14 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr8_+_52701811 | 0.14 |
ENSDART00000179692
|
bcl2l16
|
BCL2 like 16 |
| chr2_+_31671545 | 0.13 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr11_+_45153104 | 0.13 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr24_-_39701027 | 0.13 |
ENSDART00000146491
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr4_-_9891874 | 0.13 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr24_+_38201089 | 0.13 |
ENSDART00000132338
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr1_-_52497834 | 0.13 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr6_+_51713076 | 0.13 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr4_+_58667348 | 0.13 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr23_-_4704938 | 0.13 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr4_+_77933084 | 0.12 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr4_-_18775548 | 0.12 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr13_+_4671698 | 0.12 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr7_-_20103384 | 0.12 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr20_-_23439011 | 0.12 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr17_-_15600455 | 0.12 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr8_+_39511932 | 0.12 |
ENSDART00000113511
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr3_-_34816893 | 0.12 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr7_+_69019851 | 0.12 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr7_-_6458222 | 0.12 |
ENSDART00000172861
|
zgc:165555
|
zgc:165555 |
| chr6_+_48348415 | 0.11 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr13_+_12671513 | 0.11 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr19_+_37120491 | 0.11 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr11_+_44617021 | 0.11 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr4_-_33763221 | 0.11 |
ENSDART00000132613
ENSDART00000191816 ENSDART00000188473 |
si:ch211-197f20.1
|
si:ch211-197f20.1 |
| chr25_+_3549401 | 0.11 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr19_+_49721 | 0.11 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr1_-_10640748 | 0.11 |
ENSDART00000103550
|
zgc:174945
|
zgc:174945 |
| chr4_+_58661331 | 0.11 |
ENSDART00000188763
|
CR388148.2
|
|
| chr19_-_7070691 | 0.11 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr5_+_36850650 | 0.10 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr23_+_4414343 | 0.10 |
ENSDART00000081821
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr7_-_22941472 | 0.10 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr3_-_4303262 | 0.10 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr3_+_32118670 | 0.10 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr4_-_38829098 | 0.10 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr9_+_33222911 | 0.09 |
ENSDART00000135387
|
si:ch211-125e6.14
|
si:ch211-125e6.14 |
| chr19_-_7043355 | 0.09 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr11_-_27821 | 0.09 |
ENSDART00000158769
ENSDART00000172970 ENSDART00000173118 ENSDART00000168674 ENSDART00000163545 ENSDART00000173411 ENSDART00000172132 |
sp1
|
sp1 transcription factor |
| chr23_+_4414164 | 0.09 |
ENSDART00000192762
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr7_-_26262978 | 0.09 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr20_+_35445462 | 0.09 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr23_-_4705110 | 0.09 |
ENSDART00000144536
ENSDART00000129050 ENSDART00000136399 |
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr5_+_51026563 | 0.09 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr13_+_2908764 | 0.09 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr1_+_58039551 | 0.09 |
ENSDART00000073777
|
trim35-1
|
tripartite motif containing 35-1 |
| chr23_+_19759128 | 0.09 |
ENSDART00000135820
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr7_+_14005111 | 0.09 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr25_+_8407892 | 0.08 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr21_+_19525141 | 0.08 |
ENSDART00000058489
|
gzma
|
granzyme A |
| chr4_+_25181572 | 0.08 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr24_-_23998897 | 0.08 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr6_-_18976168 | 0.08 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr24_+_16985181 | 0.08 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr17_+_32623931 | 0.08 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr3_+_24708685 | 0.08 |
ENSDART00000153507
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr3_+_19245804 | 0.08 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr8_+_1766206 | 0.08 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr7_-_29534001 | 0.08 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr4_+_76973771 | 0.08 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr4_+_70015788 | 0.08 |
ENSDART00000161133
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr4_+_77948970 | 0.08 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr22_-_6229275 | 0.08 |
ENSDART00000146045
ENSDART00000179730 |
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr19_+_24488403 | 0.08 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr18_+_40354998 | 0.08 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr3_+_43086548 | 0.08 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr11_+_37905630 | 0.08 |
ENSDART00000170303
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr23_+_30967686 | 0.08 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr1_+_57176256 | 0.08 |
ENSDART00000152741
|
si:dkey-27j5.6
|
si:dkey-27j5.6 |
| chr16_+_8695595 | 0.07 |
ENSDART00000173249
|
si:cabz01093077.1
|
si:cabz01093077.1 |
| chr7_-_30926030 | 0.07 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr11_+_6752468 | 0.07 |
ENSDART00000187046
ENSDART00000182089 |
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr6_+_9867426 | 0.07 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr2_+_47945359 | 0.07 |
ENSDART00000098054
|
ftr19
|
finTRIM family, member 19 |
| chr13_-_10945288 | 0.07 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr4_+_36616118 | 0.07 |
ENSDART00000160058
ENSDART00000170506 |
si:dkey-151g22.1
|
si:dkey-151g22.1 |
| chr9_+_38481780 | 0.07 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr16_-_35586401 | 0.07 |
ENSDART00000169868
ENSDART00000166606 ENSDART00000162562 |
scmh1
|
Scm polycomb group protein homolog 1 |
| chr8_+_50190742 | 0.07 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr4_-_4261673 | 0.07 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr4_+_18960914 | 0.07 |
ENSDART00000139730
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr12_+_18663154 | 0.07 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr9_-_41817712 | 0.07 |
ENSDART00000182657
|
si:dkeyp-30e7.2
|
si:dkeyp-30e7.2 |
| chr4_+_29620959 | 0.07 |
ENSDART00000133110
|
si:ch211-214c20.1
|
si:ch211-214c20.1 |
| chr18_-_41160975 | 0.07 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr25_-_31907590 | 0.07 |
ENSDART00000149707
ENSDART00000149471 |
eif3ja
|
eukaryotic translation initiation factor 3, subunit Ja |
| chr15_+_21202820 | 0.07 |
ENSDART00000154036
|
si:dkey-52d15.2
|
si:dkey-52d15.2 |
| chr23_+_19655301 | 0.07 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr24_+_24726956 | 0.07 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr9_+_46745348 | 0.07 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr9_-_9282519 | 0.07 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr4_-_76303301 | 0.07 |
ENSDART00000158504
|
zgc:174944
|
zgc:174944 |
| chr13_+_23988442 | 0.07 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr8_+_10835456 | 0.07 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr1_+_57132140 | 0.07 |
ENSDART00000152131
|
si:ch73-94k4.3
|
si:ch73-94k4.3 |
| chr4_+_69712223 | 0.07 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr13_-_16222388 | 0.07 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr7_+_50395856 | 0.06 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr9_+_22388686 | 0.06 |
ENSDART00000182731
ENSDART00000181462 |
dgkg
|
diacylglycerol kinase, gamma |
| chr4_+_71014655 | 0.06 |
ENSDART00000170837
|
si:dkeyp-80d11.14
|
si:dkeyp-80d11.14 |
| chr20_+_6543674 | 0.06 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr24_-_39698825 | 0.06 |
ENSDART00000181089
|
CT573356.1
|
|
| chr23_-_437467 | 0.06 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr18_+_3052408 | 0.06 |
ENSDART00000181563
|
kctd14
|
potassium channel tetramerization domain containing 14 |
| chr13_+_19283936 | 0.06 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr4_+_40035116 | 0.06 |
ENSDART00000167808
|
BX649521.1
|
|
| chr1_+_56955349 | 0.06 |
ENSDART00000152758
|
si:ch211-1f22.10
|
si:ch211-1f22.10 |
| chr8_+_54013199 | 0.06 |
ENSDART00000158497
|
CABZ01079663.1
|
|
| chr18_+_7543347 | 0.06 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr4_-_43863085 | 0.06 |
ENSDART00000147130
|
si:dkey-261p22.1
|
si:dkey-261p22.1 |
| chr24_-_33366188 | 0.06 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr4_-_43616331 | 0.06 |
ENSDART00000183211
ENSDART00000150240 |
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr4_-_33073382 | 0.06 |
ENSDART00000146414
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr23_+_24989387 | 0.06 |
ENSDART00000172299
ENSDART00000145307 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr7_-_7764287 | 0.06 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr4_+_34121902 | 0.06 |
ENSDART00000170225
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr14_+_8594367 | 0.06 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr23_+_25292147 | 0.06 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr7_+_34549377 | 0.06 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr23_-_20361971 | 0.06 |
ENSDART00000133373
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr21_+_6290566 | 0.05 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr5_-_64900552 | 0.05 |
ENSDART00000073963
|
si:ch211-236k19.2
|
si:ch211-236k19.2 |
| chr25_-_3549321 | 0.05 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr4_-_39487825 | 0.05 |
ENSDART00000161855
|
si:dkey-261o4.6
|
si:dkey-261o4.6 |
| chr4_-_39186893 | 0.05 |
ENSDART00000150371
ENSDART00000121804 ENSDART00000133901 |
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr19_+_4051535 | 0.05 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosl2l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.4 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 0.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.0 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.4 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |