Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
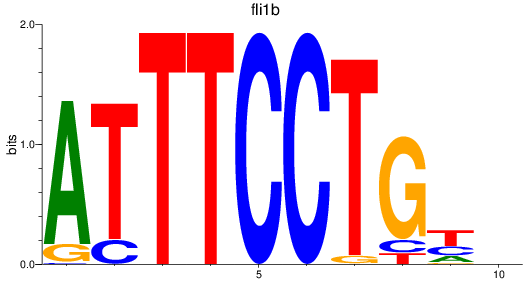
Results for fli1b
Z-value: 2.03
Transcription factors associated with fli1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fli1b
|
ENSDARG00000040080 | Fli-1 proto-oncogene, ETS transcription factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fli1b | dr11_v1_chr16_+_42018367_42018367 | 0.87 | 5.3e-02 | Click! |
Activity profile of fli1b motif
Sorted Z-values of fli1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_38285671 | 2.38 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr22_+_5687615 | 1.65 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr5_-_42272517 | 1.57 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr23_-_10175898 | 1.56 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr14_-_9128919 | 1.52 |
ENSDART00000108641
|
sh2d1ab
|
SH2 domain containing 1A duplicate b |
| chr14_-_40797117 | 1.18 |
ENSDART00000122369
|
elf1
|
E74-like ETS transcription factor 1 |
| chr9_+_2499627 | 1.18 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr19_-_19442983 | 1.11 |
ENSDART00000052649
|
sb:cb649
|
sb:cb649 |
| chr16_-_51299061 | 1.10 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr6_-_29195642 | 1.08 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr15_-_4528326 | 1.08 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr16_+_42018041 | 1.07 |
ENSDART00000134010
ENSDART00000102789 |
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr7_+_39706004 | 1.05 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr3_+_29510818 | 1.03 |
ENSDART00000055407
ENSDART00000193743 ENSDART00000123619 |
rac2
|
Rac family small GTPase 2 |
| chr17_-_37395460 | 1.00 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr1_+_1904419 | 1.00 |
ENSDART00000142874
|
si:ch211-132g1.4
|
si:ch211-132g1.4 |
| chr13_+_32144370 | 0.97 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr25_+_34014523 | 0.93 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr15_+_20239141 | 0.91 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr8_-_18010735 | 0.91 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr15_+_17756113 | 0.90 |
ENSDART00000155197
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr5_-_26879302 | 0.90 |
ENSDART00000098571
ENSDART00000139086 |
zgc:64051
|
zgc:64051 |
| chr8_-_7093507 | 0.88 |
ENSDART00000045669
|
si:dkey-222n6.2
|
si:dkey-222n6.2 |
| chr11_+_8129536 | 0.88 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr24_-_6078222 | 0.87 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr7_+_19482084 | 0.86 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr13_+_30696286 | 0.86 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr1_-_18848955 | 0.86 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr16_-_39570832 | 0.85 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr2_+_48282590 | 0.84 |
ENSDART00000035338
|
lpar5a
|
lysophosphatidic acid receptor 5a |
| chr16_-_51288178 | 0.84 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr2_-_37862380 | 0.83 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr12_-_28983584 | 0.81 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
| chr7_-_22981796 | 0.81 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr5_-_26834511 | 0.81 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr3_-_32603191 | 0.81 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr13_-_15986871 | 0.80 |
ENSDART00000189394
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr25_+_14109626 | 0.79 |
ENSDART00000109883
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr21_-_34972872 | 0.79 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr6_-_10233538 | 0.78 |
ENSDART00000182004
ENSDART00000149237 ENSDART00000148876 |
xirp2a
|
xin actin binding repeat containing 2a |
| chr9_+_54179306 | 0.78 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr16_+_35401543 | 0.78 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr8_-_4586696 | 0.77 |
ENSDART00000143511
ENSDART00000134324 |
gp1bb
|
glycoprotein Ib (platelet), beta polypeptide |
| chr19_-_2231146 | 0.77 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr8_-_31107537 | 0.75 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr18_-_16123222 | 0.74 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr21_-_28439596 | 0.74 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr7_+_20563305 | 0.73 |
ENSDART00000169661
|
si:dkey-19b23.10
|
si:dkey-19b23.10 |
| chr7_-_48263516 | 0.73 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr7_-_56606752 | 0.72 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr15_+_28410664 | 0.70 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr4_-_22310956 | 0.69 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr8_-_17997845 | 0.68 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr8_-_14209852 | 0.68 |
ENSDART00000005359
|
def6a
|
differentially expressed in FDCP 6a homolog (mouse) |
| chr5_+_37087583 | 0.67 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr19_-_15192840 | 0.67 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr11_+_24314148 | 0.67 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr4_-_22311610 | 0.67 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr15_-_2652640 | 0.67 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr9_-_33877476 | 0.67 |
ENSDART00000150035
ENSDART00000088441 ENSDART00000183210 |
si:ch73-147f11.1
|
si:ch73-147f11.1 |
| chr9_-_98982 | 0.67 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr10_+_22775253 | 0.67 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr12_-_35830625 | 0.66 |
ENSDART00000180028
|
CU459056.1
|
|
| chr16_-_10316359 | 0.66 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr21_+_25765734 | 0.66 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr3_+_24655643 | 0.65 |
ENSDART00000126148
|
zmp:0000000912
|
zmp:0000000912 |
| chr6_+_49881864 | 0.64 |
ENSDART00000075040
|
tubb1
|
tubulin, beta 1 class VI |
| chr9_+_14010823 | 0.64 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr16_-_26676685 | 0.63 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr11_-_11458208 | 0.63 |
ENSDART00000005485
|
krt93
|
keratin 93 |
| chr16_-_51271962 | 0.63 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr12_+_22576404 | 0.63 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr11_+_24313931 | 0.62 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr8_+_25342896 | 0.62 |
ENSDART00000129032
|
CR847543.1
|
|
| chr10_-_20650302 | 0.62 |
ENSDART00000142028
ENSDART00000064662 |
rassf2b
|
Ras association (RalGDS/AF-6) domain family member 2b |
| chr21_+_25198637 | 0.62 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr1_+_1915967 | 0.62 |
ENSDART00000131463
|
si:ch211-132g1.1
|
si:ch211-132g1.1 |
| chr9_+_13985567 | 0.61 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr15_-_43284021 | 0.61 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr10_+_3127809 | 0.61 |
ENSDART00000185500
|
ckmt2a
|
creatine kinase, mitochondrial 2a (sarcomeric) |
| chr3_+_59899452 | 0.61 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr5_-_34993242 | 0.60 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr9_-_31108285 | 0.60 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr19_+_43297546 | 0.60 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr15_-_551177 | 0.60 |
ENSDART00000066774
ENSDART00000154617 |
tagln
|
transgelin |
| chr7_+_17816006 | 0.59 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr16_-_51253925 | 0.59 |
ENSDART00000050644
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr18_-_40856354 | 0.59 |
ENSDART00000098865
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr17_-_12712776 | 0.59 |
ENSDART00000064511
|
il17a/f1
|
interleukin 17a/f1 |
| chr19_+_48176745 | 0.59 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr9_-_34368842 | 0.59 |
ENSDART00000140349
|
cd247l
|
CD247 antigen like |
| chr7_-_26601307 | 0.58 |
ENSDART00000188934
|
plscr3b
|
phospholipid scramblase 3b |
| chr5_-_69716501 | 0.57 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr21_-_25295087 | 0.57 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr20_+_25340814 | 0.56 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr16_+_11818126 | 0.56 |
ENSDART00000145727
|
cxcr3.2
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 2 |
| chr7_+_11543999 | 0.56 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr19_-_12404590 | 0.55 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr13_-_45523026 | 0.55 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr17_-_7733037 | 0.55 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr19_-_15192638 | 0.55 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr22_-_15280638 | 0.55 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr19_+_43716607 | 0.55 |
ENSDART00000133199
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr7_+_19762595 | 0.54 |
ENSDART00000130347
|
si:dkey-9k7.3
|
si:dkey-9k7.3 |
| chr3_+_27713610 | 0.54 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr23_+_42813415 | 0.54 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr11_+_22109887 | 0.54 |
ENSDART00000122136
|
MDFI
|
si:dkey-91m3.1 |
| chr13_-_13751017 | 0.53 |
ENSDART00000180182
|
ky
|
kyphoscoliosis peptidase |
| chr21_-_41025340 | 0.53 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr20_-_21672970 | 0.53 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr3_+_17939828 | 0.53 |
ENSDART00000185047
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr17_-_2039511 | 0.52 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr8_+_3379815 | 0.52 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr8_-_19975087 | 0.52 |
ENSDART00000182220
|
lpxn
|
leupaxin |
| chr8_+_20398445 | 0.52 |
ENSDART00000134755
|
zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr5_+_22133153 | 0.52 |
ENSDART00000016214
|
msna
|
moesin a |
| chr9_+_13999620 | 0.52 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr17_+_33453689 | 0.51 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr24_-_31904924 | 0.51 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr1_-_29658721 | 0.51 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr13_-_42749916 | 0.51 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr22_-_17729778 | 0.51 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr21_+_30306369 | 0.50 |
ENSDART00000145050
ENSDART00000059420 |
lcp2a
|
lymphocyte cytosolic protein 2a |
| chr2_+_22495274 | 0.50 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr22_-_18164671 | 0.50 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr10_-_11012000 | 0.50 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr14_+_34501245 | 0.50 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr18_+_49969568 | 0.50 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr1_-_52498146 | 0.50 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr5_+_30741730 | 0.50 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr9_-_30243393 | 0.50 |
ENSDART00000089539
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr10_+_9553935 | 0.49 |
ENSDART00000028855
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr3_+_17933132 | 0.49 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr8_+_48613040 | 0.49 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr16_-_23797570 | 0.49 |
ENSDART00000077834
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr13_-_37653840 | 0.48 |
ENSDART00000143806
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr22_-_31672566 | 0.48 |
ENSDART00000128247
|
lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr14_-_33613794 | 0.48 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr22_-_17652112 | 0.48 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr9_+_23772516 | 0.48 |
ENSDART00000183126
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr19_+_43579786 | 0.48 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr16_+_42018367 | 0.47 |
ENSDART00000058613
|
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr21_+_22985078 | 0.47 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr12_+_23892972 | 0.47 |
ENSDART00000152852
|
svila
|
supervillin a |
| chr25_+_35553542 | 0.47 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr14_-_897874 | 0.47 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr10_+_20113830 | 0.47 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr16_-_9802449 | 0.47 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr1_-_45157243 | 0.46 |
ENSDART00000131882
|
mucms1
|
mucin, multiple PTS and SEA group, member 1 |
| chr19_-_325584 | 0.46 |
ENSDART00000134266
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr7_+_18017756 | 0.46 |
ENSDART00000173717
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr12_+_27536270 | 0.45 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr5_+_46424437 | 0.45 |
ENSDART00000186511
|
vcana
|
versican a |
| chr25_-_16589461 | 0.45 |
ENSDART00000064204
|
cpa4
|
carboxypeptidase A4 |
| chr6_-_55423220 | 0.44 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr8_-_25814263 | 0.44 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr7_-_32659048 | 0.44 |
ENSDART00000093279
|
spi1b
|
Spi-1 proto-oncogene b |
| chr4_-_11064073 | 0.44 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr3_-_21382491 | 0.44 |
ENSDART00000016562
|
itga2b
|
integrin, alpha 2b |
| chr2_-_41964402 | 0.44 |
ENSDART00000131278
|
si:dkey-97a13.6
|
si:dkey-97a13.6 |
| chr7_-_32659345 | 0.44 |
ENSDART00000126831
ENSDART00000036729 |
spi1b
|
Spi-1 proto-oncogene b |
| chr25_-_16600811 | 0.44 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr3_+_24612191 | 0.43 |
ENSDART00000190998
|
zgc:171506
|
zgc:171506 |
| chr24_-_2423791 | 0.43 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr21_+_30794351 | 0.43 |
ENSDART00000139486
|
zgc:158225
|
zgc:158225 |
| chr6_-_8480815 | 0.43 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr2_-_25140022 | 0.42 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr9_-_9419704 | 0.42 |
ENSDART00000138996
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr15_+_17251191 | 0.42 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr23_+_44049509 | 0.42 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr21_-_11632403 | 0.42 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr1_+_33558555 | 0.42 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr21_+_261490 | 0.42 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr3_+_24641489 | 0.42 |
ENSDART00000055590
ENSDART00000186635 |
zgc:113411
|
zgc:113411 |
| chr18_+_38288877 | 0.41 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr14_+_23709543 | 0.41 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_-_8653385 | 0.41 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr10_+_7798087 | 0.41 |
ENSDART00000141824
|
sftpba
|
surfactant protein Ba |
| chr22_-_18164835 | 0.41 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr12_+_20552190 | 0.41 |
ENSDART00000113224
|
gna13a
|
guanine nucleotide binding protein (G protein), alpha 13a |
| chr9_+_21165484 | 0.41 |
ENSDART00000177286
|
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr20_+_15600167 | 0.41 |
ENSDART00000171991
|
faslg
|
Fas ligand (TNF superfamily, member 6) |
| chr13_-_37189560 | 0.41 |
ENSDART00000134789
|
si:dkeyp-77c8.2
|
si:dkeyp-77c8.2 |
| chr9_-_51563575 | 0.40 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr3_+_22377312 | 0.40 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr1_+_1896737 | 0.40 |
ENSDART00000152442
|
si:ch211-132g1.6
|
si:ch211-132g1.6 |
| chr2_+_13069168 | 0.40 |
ENSDART00000192832
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr13_-_7031033 | 0.40 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr3_+_3545825 | 0.40 |
ENSDART00000109060
|
CR589947.1
|
|
| chr25_-_21031007 | 0.40 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr1_-_52497834 | 0.40 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr5_+_42136359 | 0.39 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr14_-_34059681 | 0.39 |
ENSDART00000003993
|
itk
|
IL2 inducible T cell kinase |
| chr21_+_29077509 | 0.39 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr23_+_7505943 | 0.39 |
ENSDART00000135787
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr10_+_36197968 | 0.39 |
ENSDART00000114102
|
COA4
|
si:ch211-215a9.5 |
| chr13_+_24402406 | 0.39 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr18_-_20869175 | 0.38 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr24_-_40726073 | 0.38 |
ENSDART00000168100
|
smyhc2
|
slow myosin heavy chain 2 |
| chr18_+_27598755 | 0.38 |
ENSDART00000193808
|
cd82b
|
CD82 molecule b |
Network of associatons between targets according to the STRING database.
First level regulatory network of fli1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.3 | 1.0 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.3 | 1.4 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.3 | 0.8 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 1.3 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.7 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.2 | 1.0 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.2 | 0.6 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.2 | 0.6 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 0.9 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.5 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.2 | 0.5 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.4 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.4 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.5 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 1.8 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.7 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 3.4 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 0.3 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.3 | GO:0045123 | cellular extravasation(GO:0045123) |
| 0.1 | 0.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.3 | GO:0002637 | regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.1 | 0.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.5 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.3 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 0.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.4 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.2 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.3 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 1.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.8 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.8 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.6 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 1.1 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 1.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.5 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 2.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.6 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.9 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.1 | GO:1990120 | lymphoid progenitor cell differentiation(GO:0002320) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of blood vessel endothelial cell migration(GO:0043536) regulation of cell migration involved in sprouting angiogenesis(GO:0090049) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.5 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.0 | 0.3 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 1.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.9 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.7 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.2 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 1.5 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 1.8 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.4 | GO:0098703 | response to heat(GO:0009408) calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.7 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.0 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0071962 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.4 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 2.2 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.0 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.8 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.3 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.0 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 1.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.9 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 0.6 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.2 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 4.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.6 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.0 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 0.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 1.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.7 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 0.6 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.2 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 1.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.5 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 3.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.2 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.4 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.1 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 1.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.1 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 4.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 1.1 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 7.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |