Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
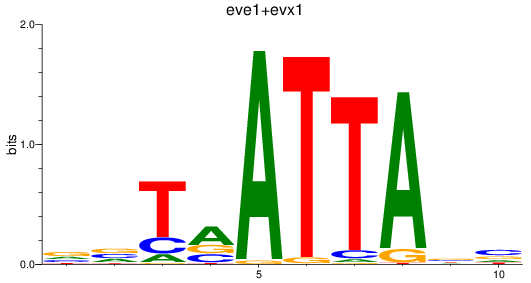
Results for eve1+evx1
Z-value: 0.73
Transcription factors associated with eve1+evx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
eve1
|
ENSDARG00000056012 | even-skipped-like1 |
|
evx1
|
ENSDARG00000099365 | even-skipped homeobox 1 |
|
evx1
|
ENSDARG00000100087 | even-skipped homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| evx1 | dr11_v1_chr19_-_19871211_19871211 | 0.09 | 8.8e-01 | Click! |
Activity profile of eve1+evx1 motif
Sorted Z-values of eve1+evx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_2414063 | 0.86 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr20_-_23426339 | 0.69 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr21_+_25777425 | 0.69 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr25_+_3327071 | 0.57 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr3_+_1005059 | 0.54 |
ENSDART00000132460
|
zgc:172051
|
zgc:172051 |
| chr16_-_54455573 | 0.48 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr10_+_21867307 | 0.46 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr7_+_69019851 | 0.41 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr9_-_35633827 | 0.39 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr10_+_17714866 | 0.36 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr2_+_6253246 | 0.33 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr3_+_17537352 | 0.31 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr10_-_21362071 | 0.30 |
ENSDART00000125167
|
avd
|
avidin |
| chr7_-_27685365 | 0.27 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr19_-_18313303 | 0.27 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr2_-_50225411 | 0.26 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr20_-_40755614 | 0.26 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr10_-_21362320 | 0.25 |
ENSDART00000189789
|
avd
|
avidin |
| chr19_+_31585917 | 0.24 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr16_-_42056137 | 0.23 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr20_+_41021054 | 0.20 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr25_-_11378623 | 0.19 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr10_+_7709724 | 0.18 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr12_-_19007834 | 0.16 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr2_-_57076687 | 0.16 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr10_-_35257458 | 0.16 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr6_-_40352215 | 0.15 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr16_-_29458806 | 0.14 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr2_-_32826108 | 0.14 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr2_-_32825917 | 0.13 |
ENSDART00000180409
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr21_-_44772738 | 0.13 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr2_-_21819421 | 0.12 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr9_-_53666031 | 0.12 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr11_+_31864921 | 0.12 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr23_-_31913231 | 0.12 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr21_-_19919020 | 0.11 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr17_+_16046132 | 0.11 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr23_+_7548797 | 0.10 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr3_-_26183699 | 0.10 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr6_+_41191482 | 0.10 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr10_-_44560165 | 0.09 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr1_-_9195629 | 0.09 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr19_+_40856807 | 0.09 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr3_+_18398876 | 0.09 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr1_-_58505626 | 0.09 |
ENSDART00000171304
|
si:ch73-236c18.8
|
si:ch73-236c18.8 |
| chr3_-_16719244 | 0.09 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr19_+_40856534 | 0.09 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr16_+_5774977 | 0.09 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr10_+_25726694 | 0.09 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr5_+_64732270 | 0.09 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr6_+_28208973 | 0.08 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr21_+_30355767 | 0.08 |
ENSDART00000189948
|
CR749164.1
|
|
| chr8_+_53423408 | 0.08 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr10_+_36345176 | 0.08 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr17_-_23446760 | 0.08 |
ENSDART00000104715
|
pcgf5a
|
polycomb group ring finger 5a |
| chr16_+_17389116 | 0.08 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr3_+_54047342 | 0.08 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr2_-_11258547 | 0.08 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr21_+_1381276 | 0.08 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr21_+_43404945 | 0.08 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr25_-_25384045 | 0.08 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr23_+_40460333 | 0.08 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr22_+_17205608 | 0.07 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr21_+_10773522 | 0.07 |
ENSDART00000143257
|
znf532
|
zinc finger protein 532 |
| chr11_+_35171406 | 0.07 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr21_-_27185915 | 0.07 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr4_+_12612723 | 0.07 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr11_+_45323455 | 0.07 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr12_-_4408828 | 0.07 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr18_+_9615147 | 0.07 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr23_+_31913292 | 0.07 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr25_+_14507567 | 0.07 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr20_-_13722514 | 0.07 |
ENSDART00000152458
|
si:ch211-122h15.4
|
si:ch211-122h15.4 |
| chr20_+_37294112 | 0.07 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr9_-_24031461 | 0.07 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr17_-_28198099 | 0.07 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr10_-_44482911 | 0.07 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr9_-_19161982 | 0.07 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr25_+_35891342 | 0.07 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr9_+_13120419 | 0.06 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr24_+_1023839 | 0.06 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr23_-_33361425 | 0.06 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr22_-_12746539 | 0.06 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr16_+_31804590 | 0.06 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr21_+_34814444 | 0.06 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr11_-_44876005 | 0.06 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr8_+_2438638 | 0.06 |
ENSDART00000141263
|
ENKD1
|
si:ch211-220d9.3 |
| chr1_+_16397063 | 0.06 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr22_-_27709692 | 0.06 |
ENSDART00000172458
|
CR547131.1
|
|
| chr21_-_5205617 | 0.06 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr2_-_16380283 | 0.06 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr17_+_21295132 | 0.06 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr23_+_4646194 | 0.06 |
ENSDART00000092344
|
LO017700.1
|
|
| chr7_-_17591007 | 0.06 |
ENSDART00000171023
|
CU672228.1
|
|
| chr12_+_46582727 | 0.05 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr10_+_518546 | 0.05 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr13_+_38814521 | 0.05 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr23_-_4966821 | 0.05 |
ENSDART00000147122
ENSDART00000145760 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr4_+_12612145 | 0.05 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr4_-_71912457 | 0.05 |
ENSDART00000179685
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr15_+_9053059 | 0.05 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr5_+_19314574 | 0.05 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr15_+_22014029 | 0.05 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr4_+_11384891 | 0.05 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr6_-_55585423 | 0.05 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr2_+_30464829 | 0.05 |
ENSDART00000181432
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr25_+_26923193 | 0.05 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr19_-_657439 | 0.05 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr1_+_16396870 | 0.05 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr10_-_17587832 | 0.05 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr1_-_47071979 | 0.05 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr12_-_32426685 | 0.05 |
ENSDART00000105574
|
enpp7.2
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 2 |
| chr15_-_22074315 | 0.05 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr5_+_26765275 | 0.05 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr19_+_15443540 | 0.05 |
ENSDART00000193355
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr21_+_40292801 | 0.05 |
ENSDART00000174220
|
si:ch211-218m3.16
|
si:ch211-218m3.16 |
| chr2_+_31492662 | 0.05 |
ENSDART00000123495
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr23_+_31912882 | 0.05 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr5_+_27137473 | 0.04 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr10_+_41765944 | 0.04 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr8_-_24252933 | 0.04 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr2_-_10896745 | 0.04 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr19_+_5543072 | 0.04 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr4_+_52356485 | 0.04 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr8_-_39822917 | 0.04 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr4_+_13586689 | 0.04 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr6_+_9870192 | 0.04 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr21_-_3853204 | 0.04 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_+_30465102 | 0.04 |
ENSDART00000188404
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr22_-_37686966 | 0.04 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_+_306036 | 0.04 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr2_-_1443692 | 0.04 |
ENSDART00000004533
|
rpe65a
|
retinal pigment epithelium-specific protein 65a |
| chr21_+_26390549 | 0.04 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr18_+_50461981 | 0.03 |
ENSDART00000158761
|
CU896640.1
|
|
| chr1_+_58116900 | 0.03 |
ENSDART00000160436
|
si:ch211-15j1.5
|
si:ch211-15j1.5 |
| chr15_+_5360407 | 0.03 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr1_+_55758257 | 0.03 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr2_+_56213694 | 0.03 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr24_+_12894282 | 0.03 |
ENSDART00000061301
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr5_+_64732036 | 0.03 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr21_+_11244068 | 0.03 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr21_-_18993110 | 0.03 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr21_-_26490186 | 0.03 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr15_+_841383 | 0.03 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr12_+_46582168 | 0.03 |
ENSDART00000189402
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr7_-_39537292 | 0.03 |
ENSDART00000173965
|
otog
|
otogelin |
| chr8_-_31107537 | 0.03 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr11_+_24703108 | 0.03 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr7_-_17816478 | 0.03 |
ENSDART00000149403
|
ecsit
|
ECSIT signalling integrator |
| chr23_+_26946744 | 0.03 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr1_+_51191049 | 0.03 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr18_-_1185772 | 0.03 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr23_+_44374041 | 0.02 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr1_-_17715493 | 0.02 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr3_-_8399774 | 0.02 |
ENSDART00000190238
ENSDART00000113140 ENSDART00000132949 |
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr9_-_202805 | 0.02 |
ENSDART00000182260
|
CABZ01067557.1
|
|
| chr14_-_8625845 | 0.02 |
ENSDART00000054760
|
zgc:162144
|
zgc:162144 |
| chr10_+_16036246 | 0.02 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr23_-_19230627 | 0.02 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr10_-_11385155 | 0.02 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr1_-_10647484 | 0.02 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr17_+_10578823 | 0.01 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr6_+_515181 | 0.01 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr23_+_33963619 | 0.01 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr1_+_27023687 | 0.01 |
ENSDART00000189093
|
bnc2
|
basonuclin 2 |
| chr18_+_28106139 | 0.01 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr5_+_23242370 | 0.01 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr23_-_31913069 | 0.01 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr21_-_31013817 | 0.01 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr6_-_6487876 | 0.01 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr7_-_17816175 | 0.01 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr2_+_32826235 | 0.01 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr11_+_42726712 | 0.01 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr23_-_20051369 | 0.01 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr16_+_35661331 | 0.01 |
ENSDART00000161725
|
map7d1a
|
MAP7 domain containing 1a |
| chr7_+_30787903 | 0.01 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr7_-_64589920 | 0.01 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr15_-_37835638 | 0.01 |
ENSDART00000128922
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr16_-_21038015 | 0.01 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr1_+_27024068 | 0.01 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr7_-_40145097 | 0.01 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr8_-_27656765 | 0.00 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr17_-_23616626 | 0.00 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr24_-_25004553 | 0.00 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr8_+_52503340 | 0.00 |
ENSDART00000168838
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr18_-_12052132 | 0.00 |
ENSDART00000074361
|
zgc:110789
|
zgc:110789 |
| chr8_-_15129573 | 0.00 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr5_+_29794058 | 0.00 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr16_-_25829779 | 0.00 |
ENSDART00000086301
|
irge4
|
immunity-related GTPase family, e4 |
| chr19_-_20430892 | 0.00 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of eve1+evx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.6 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.3 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.0 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0009217 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.2 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |