Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
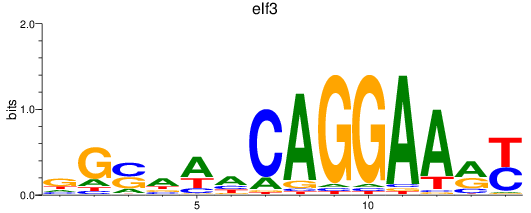
Results for elf3
Z-value: 1.05
Transcription factors associated with elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
elf3
|
ENSDARG00000077982 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| elf3 | dr11_v1_chr22_+_661711_661711 | 0.85 | 6.5e-02 | Click! |
Activity profile of elf3 motif
Sorted Z-values of elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_1904419 | 0.62 |
ENSDART00000142874
|
si:ch211-132g1.4
|
si:ch211-132g1.4 |
| chr20_+_38285671 | 0.57 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr22_+_20710434 | 0.56 |
ENSDART00000135521
|
peak3
|
PEAK family member 3 |
| chr23_-_10175898 | 0.54 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr15_+_934660 | 0.54 |
ENSDART00000154248
|
si:dkey-77f5.10
|
si:dkey-77f5.10 |
| chr9_-_33081781 | 0.51 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr11_-_41084358 | 0.49 |
ENSDART00000166999
|
zgc:175088
|
zgc:175088 |
| chr1_+_1896737 | 0.47 |
ENSDART00000152442
|
si:ch211-132g1.6
|
si:ch211-132g1.6 |
| chr7_+_35075847 | 0.46 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr3_+_29942338 | 0.44 |
ENSDART00000158220
|
ifi35
|
interferon-induced protein 35 |
| chr5_-_29531948 | 0.42 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr15_-_2652640 | 0.42 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr13_+_30696286 | 0.42 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr17_-_24890843 | 0.42 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr3_+_24612191 | 0.41 |
ENSDART00000190998
|
zgc:171506
|
zgc:171506 |
| chr1_-_18848955 | 0.41 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr10_+_38417512 | 0.41 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr2_-_36918709 | 0.40 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr9_-_54304684 | 0.40 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr20_+_19176959 | 0.38 |
ENSDART00000163276
ENSDART00000020099 |
blk
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr15_-_34892664 | 0.37 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr5_-_54712159 | 0.37 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr16_+_38201840 | 0.36 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr7_-_22981796 | 0.35 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr21_-_22673758 | 0.35 |
ENSDART00000164910
|
gig2i
|
grass carp reovirus (GCRV)-induced gene 2i |
| chr5_-_37959874 | 0.34 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr5_+_37087583 | 0.34 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr19_-_12404590 | 0.33 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr17_-_21368775 | 0.31 |
ENSDART00000058027
|
shtn1
|
shootin 1 |
| chr11_+_20899029 | 0.30 |
ENSDART00000163029
|
zgc:162182
|
zgc:162182 |
| chr24_+_37449893 | 0.30 |
ENSDART00000164405
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr15_+_1534644 | 0.30 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr6_-_8489810 | 0.29 |
ENSDART00000124643
|
rasal3
|
RAS protein activator like 3 |
| chr24_+_6353394 | 0.29 |
ENSDART00000165118
|
CR352329.1
|
|
| chr22_+_18886209 | 0.29 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr20_-_22798794 | 0.29 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr6_+_28752943 | 0.29 |
ENSDART00000078447
ENSDART00000146574 |
tprg1
|
tumor protein p63 regulated 1 |
| chr21_-_22676323 | 0.28 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr8_-_52715911 | 0.27 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr19_-_40199081 | 0.27 |
ENSDART00000051970
ENSDART00000151079 |
grn2
|
granulin 2 |
| chr7_-_18691637 | 0.26 |
ENSDART00000183715
|
si:ch211-119e14.9
|
si:ch211-119e14.9 |
| chr12_-_44043285 | 0.26 |
ENSDART00000163074
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr22_+_1170294 | 0.26 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr8_-_39838660 | 0.26 |
ENSDART00000139266
|
ftr98
|
finTRIM family, member 98 |
| chr5_-_29152457 | 0.26 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr20_+_13533544 | 0.25 |
ENSDART00000143115
|
sytl3
|
synaptotagmin-like 3 |
| chr13_-_33700461 | 0.25 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr6_-_442163 | 0.25 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr21_+_25198637 | 0.24 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr23_+_36087219 | 0.24 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr1_-_29658721 | 0.24 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr6_+_18298444 | 0.24 |
ENSDART00000166018
|
card14
|
caspase recruitment domain family, member 14 |
| chr2_+_36109002 | 0.23 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr20_-_2619316 | 0.23 |
ENSDART00000185777
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr20_+_54290356 | 0.23 |
ENSDART00000173347
|
zp2.2
|
zona pellucida glycoprotein 2, tandem duplicate 2 |
| chr15_-_951118 | 0.23 |
ENSDART00000171427
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr11_-_21304452 | 0.23 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr6_-_55254786 | 0.22 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr19_+_43341115 | 0.22 |
ENSDART00000145846
ENSDART00000102384 |
sesn2
|
sestrin 2 |
| chr20_+_13141408 | 0.22 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr25_+_13205878 | 0.22 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr22_-_17652112 | 0.22 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr19_+_48176745 | 0.21 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr16_+_29514473 | 0.21 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr1_+_532766 | 0.21 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr19_+_43341424 | 0.21 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr20_-_13625588 | 0.21 |
ENSDART00000078893
|
sytl3
|
synaptotagmin-like 3 |
| chr22_-_17652914 | 0.21 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr7_+_29509255 | 0.21 |
ENSDART00000076172
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr5_-_26834511 | 0.20 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr24_+_39990695 | 0.20 |
ENSDART00000040281
|
BX323854.1
|
|
| chr4_-_58964138 | 0.20 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr4_-_22310956 | 0.20 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr1_-_8020589 | 0.20 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr3_+_49576662 | 0.19 |
ENSDART00000162275
|
CR394535.1
|
|
| chr13_+_31070181 | 0.19 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr14_-_11529311 | 0.19 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr9_+_2499627 | 0.19 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr13_-_21672131 | 0.18 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr4_-_4261673 | 0.18 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr20_+_34770197 | 0.18 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr1_-_59252973 | 0.18 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr19_+_4038589 | 0.18 |
ENSDART00000169271
|
btr23
|
bloodthirsty-related gene family, member 23 |
| chr11_+_8129536 | 0.18 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr15_-_28677725 | 0.18 |
ENSDART00000060255
|
blmh
|
bleomycin hydrolase |
| chr7_+_22981909 | 0.18 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr5_+_32141790 | 0.17 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr7_+_26545502 | 0.17 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr1_-_11104805 | 0.17 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr15_+_15415623 | 0.16 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr21_+_6114305 | 0.16 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr4_+_7822773 | 0.16 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr4_-_992063 | 0.16 |
ENSDART00000181630
ENSDART00000183898 ENSDART00000160902 |
naga
|
N-acetylgalactosaminidase, alpha |
| chr21_+_35296246 | 0.16 |
ENSDART00000076750
|
il12bb
|
interleukin 12B, b |
| chr24_-_6038025 | 0.15 |
ENSDART00000077819
ENSDART00000139216 |
ftr61
|
finTRIM family, member 61 |
| chr8_-_36399884 | 0.15 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr2_-_59205393 | 0.15 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr1_-_59407322 | 0.15 |
ENSDART00000161661
|
si:ch211-188p14.5
|
si:ch211-188p14.5 |
| chr16_-_15387459 | 0.15 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr4_-_5795309 | 0.15 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr1_-_21881818 | 0.15 |
ENSDART00000047728
|
melk
|
maternal embryonic leucine zipper kinase |
| chr2_-_59303338 | 0.15 |
ENSDART00000100987
ENSDART00000140840 |
ftr35
|
finTRIM family, member 35 |
| chr8_-_42594380 | 0.15 |
ENSDART00000140126
ENSDART00000135238 ENSDART00000192764 |
dok2
|
docking protein 2 |
| chr9_+_3153717 | 0.15 |
ENSDART00000191958
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr3_-_10706846 | 0.14 |
ENSDART00000157260
|
si:ch73-1f23.1
|
si:ch73-1f23.1 |
| chr22_-_24967348 | 0.14 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr14_-_897874 | 0.14 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr10_-_1523253 | 0.14 |
ENSDART00000179510
ENSDART00000176548 ENSDART00000180368 ENSDART00000185270 |
WDR70
|
WD repeat domain 70 |
| chr7_+_22981441 | 0.14 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr5_+_24245682 | 0.14 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr13_-_34858500 | 0.13 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr20_-_18382708 | 0.13 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr13_-_35908275 | 0.13 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr4_-_7869731 | 0.13 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr9_+_48108174 | 0.13 |
ENSDART00000077260
|
cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr15_+_3808996 | 0.13 |
ENSDART00000110227
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr8_+_22438398 | 0.13 |
ENSDART00000135145
|
zgc:153759
|
zgc:153759 |
| chr23_+_19790962 | 0.13 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr7_+_19762595 | 0.13 |
ENSDART00000130347
|
si:dkey-9k7.3
|
si:dkey-9k7.3 |
| chr11_-_21303946 | 0.12 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr9_+_3153466 | 0.12 |
ENSDART00000135619
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr8_-_7093507 | 0.12 |
ENSDART00000045669
|
si:dkey-222n6.2
|
si:dkey-222n6.2 |
| chr22_-_30973791 | 0.12 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr13_-_35907768 | 0.12 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr3_+_43826018 | 0.12 |
ENSDART00000166197
ENSDART00000165145 |
litaf
|
lipopolysaccharide-induced TNF factor |
| chr24_-_13349464 | 0.12 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr10_-_41450367 | 0.12 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr8_+_25342896 | 0.12 |
ENSDART00000129032
|
CR847543.1
|
|
| chr14_+_41318412 | 0.12 |
ENSDART00000064614
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr13_+_1089942 | 0.12 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr22_+_2598998 | 0.12 |
ENSDART00000176665
|
CU570894.1
|
|
| chr23_-_19103855 | 0.12 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr23_+_32101361 | 0.12 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr14_+_6287871 | 0.12 |
ENSDART00000105344
|
tbc1d2
|
TBC1 domain family, member 2 |
| chr2_+_36646451 | 0.12 |
ENSDART00000039174
|
klhl6
|
kelch-like family member 6 |
| chr1_+_33558555 | 0.12 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr14_+_41318604 | 0.11 |
ENSDART00000167042
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr4_+_70414733 | 0.11 |
ENSDART00000162778
|
si:dkey-190j3.3
|
si:dkey-190j3.3 |
| chr1_-_24349759 | 0.11 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr21_+_14511685 | 0.11 |
ENSDART00000102099
|
CR392036.1
|
|
| chr21_-_45073764 | 0.11 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr24_-_38113208 | 0.11 |
ENSDART00000079003
|
crp
|
c-reactive protein, pentraxin-related |
| chr8_+_20157798 | 0.11 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr9_+_34952203 | 0.11 |
ENSDART00000121828
ENSDART00000142347 |
tfdp1a
|
transcription factor Dp-1, a |
| chr5_-_23464970 | 0.11 |
ENSDART00000141028
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr14_+_20911310 | 0.11 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr9_+_3282369 | 0.11 |
ENSDART00000044128
|
hat1
|
histone acetyltransferase 1 |
| chr5_+_42136359 | 0.11 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr3_-_36210344 | 0.11 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr23_+_32101202 | 0.11 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr15_-_2734560 | 0.11 |
ENSDART00000153853
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr3_+_19245804 | 0.11 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr21_-_40173821 | 0.11 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr22_+_16022211 | 0.10 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr7_-_24204665 | 0.10 |
ENSDART00000167141
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr19_-_3741602 | 0.10 |
ENSDART00000170301
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr12_-_33558879 | 0.10 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr16_-_17300030 | 0.10 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr12_-_33558727 | 0.10 |
ENSDART00000086087
|
mbtd1
|
mbt domain containing 1 |
| chr10_+_42169982 | 0.10 |
ENSDART00000190905
|
CU467905.1
|
|
| chr5_+_40835601 | 0.10 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr1_+_12301913 | 0.10 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr23_-_4705110 | 0.10 |
ENSDART00000144536
ENSDART00000129050 ENSDART00000136399 |
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr3_+_57997980 | 0.10 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr4_+_35590588 | 0.10 |
ENSDART00000183582
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr5_-_29534748 | 0.10 |
ENSDART00000159587
|
AL831768.1
|
|
| chr4_-_12723585 | 0.10 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr2_-_59265521 | 0.10 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr17_+_44441042 | 0.09 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr1_-_34450784 | 0.09 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr2_-_59376399 | 0.09 |
ENSDART00000137134
|
ftr38
|
finTRIM family, member 38 |
| chr11_+_13423776 | 0.09 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr21_+_21244046 | 0.09 |
ENSDART00000058306
ENSDART00000189665 ENSDART00000131890 |
lifrb
|
leukemia inhibitory factor receptor alpha b |
| chr10_-_105100 | 0.09 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr4_+_37936839 | 0.09 |
ENSDART00000164505
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr15_+_14854666 | 0.09 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr21_+_6197223 | 0.09 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr5_+_37406358 | 0.09 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr2_-_59327299 | 0.09 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr1_+_11659861 | 0.08 |
ENSDART00000054787
|
BX569798.1
|
|
| chr7_+_73295890 | 0.08 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr24_-_13349802 | 0.08 |
ENSDART00000164729
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr16_-_40426837 | 0.08 |
ENSDART00000193690
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr19_+_47476908 | 0.08 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr25_-_34632050 | 0.08 |
ENSDART00000170671
|
megf8
|
multiple EGF-like-domains 8 |
| chr25_+_34749812 | 0.08 |
ENSDART00000185712
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr15_-_35100858 | 0.08 |
ENSDART00000099677
|
bloc1s3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr17_-_44440832 | 0.08 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr16_+_43368572 | 0.08 |
ENSDART00000032778
ENSDART00000193897 |
rnf144b
|
ring finger protein 144B |
| chr11_+_36665359 | 0.08 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr3_+_52737565 | 0.08 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr2_-_24068848 | 0.08 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr25_+_5015019 | 0.07 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr8_-_13029297 | 0.07 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr13_+_10023256 | 0.07 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr24_+_20916751 | 0.07 |
ENSDART00000043193
|
cst14a.1
|
cystatin 14a, tandem duplicate 1 |
| chr7_-_19915414 | 0.07 |
ENSDART00000144180
|
zgc:110591
|
zgc:110591 |
| chr2_+_36721357 | 0.07 |
ENSDART00000019063
|
pdcd10b
|
programmed cell death 10b |
| chr1_+_57774454 | 0.07 |
ENSDART00000152507
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr1_+_58400409 | 0.07 |
ENSDART00000111526
|
si:ch73-59p9.2
|
si:ch73-59p9.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 0.4 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.1 | 0.4 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.2 | GO:0045226 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.0 | 0.2 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.4 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0051916 | C-X-C chemokine receptor activity(GO:0016494) granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |