Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
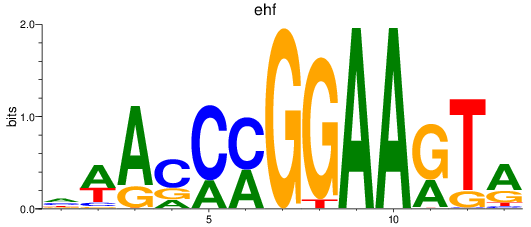
Results for ehf
Z-value: 1.30
Transcription factors associated with ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ehf
|
ENSDARG00000052115 | ets homologous factor |
|
ehf
|
ENSDARG00000112589 | ets homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ehf | dr11_v1_chr25_+_10410620_10410620 | 0.93 | 2.1e-02 | Click! |
Activity profile of ehf motif
Sorted Z-values of ehf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_33081978 | 1.28 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr9_-_33081781 | 1.27 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr1_-_45177373 | 0.92 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr15_-_29598679 | 0.85 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr7_+_45975537 | 0.78 |
ENSDART00000170253
|
plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr6_-_49063085 | 0.72 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr15_-_29598444 | 0.66 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr12_+_38878830 | 0.63 |
ENSDART00000156926
|
si:ch211-39f2.3
|
si:ch211-39f2.3 |
| chr24_-_6078222 | 0.61 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr1_-_69444 | 0.59 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr7_-_19923249 | 0.59 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr11_+_8129536 | 0.55 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr16_-_31756859 | 0.54 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr10_+_26667475 | 0.54 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr5_-_30615901 | 0.54 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr15_+_934660 | 0.53 |
ENSDART00000154248
|
si:dkey-77f5.10
|
si:dkey-77f5.10 |
| chr15_-_2652640 | 0.52 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr18_+_22994427 | 0.52 |
ENSDART00000173131
ENSDART00000172995 ENSDART00000172923 ENSDART00000141521 ENSDART00000173201 ENSDART00000059961 |
cbfb
|
core-binding factor, beta subunit |
| chr21_+_25765734 | 0.52 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr4_-_22311610 | 0.52 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr22_+_5687615 | 0.50 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr22_+_19220459 | 0.50 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr18_+_22994113 | 0.49 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr12_+_38929663 | 0.49 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr7_-_20103384 | 0.48 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr3_+_27713610 | 0.47 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr20_-_3238110 | 0.47 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr17_-_37395460 | 0.47 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr1_-_8020589 | 0.45 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr19_-_32493866 | 0.43 |
ENSDART00000052090
|
fuca1.2
|
alpha-L-fucosidase 1, tandem duplicate 2 |
| chr22_-_17653143 | 0.43 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr3_+_34120191 | 0.43 |
ENSDART00000020017
ENSDART00000151700 |
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr13_+_24402406 | 0.42 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr6_-_442163 | 0.41 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr7_+_19374683 | 0.39 |
ENSDART00000162700
|
snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr3_+_40809011 | 0.39 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr21_-_7035599 | 0.39 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr11_+_11719575 | 0.39 |
ENSDART00000003891
|
jupa
|
junction plakoglobin a |
| chr24_+_6353394 | 0.38 |
ENSDART00000165118
|
CR352329.1
|
|
| chr7_-_6754012 | 0.38 |
ENSDART00000113658
|
zgc:55262
|
zgc:55262 |
| chr8_-_39838660 | 0.38 |
ENSDART00000139266
|
ftr98
|
finTRIM family, member 98 |
| chr3_+_28581397 | 0.38 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr20_+_13533544 | 0.38 |
ENSDART00000143115
|
sytl3
|
synaptotagmin-like 3 |
| chr8_-_31107537 | 0.37 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr9_+_33417969 | 0.37 |
ENSDART00000024795
|
gpr34b
|
G protein-coupled receptor 34b |
| chr5_+_22459087 | 0.37 |
ENSDART00000134781
|
BX546499.1
|
|
| chr18_-_40856354 | 0.36 |
ENSDART00000098865
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr21_-_41025340 | 0.36 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr10_+_9553935 | 0.36 |
ENSDART00000028855
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr10_-_7785930 | 0.36 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr17_-_50311009 | 0.35 |
ENSDART00000153653
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr1_+_33558555 | 0.35 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr20_-_34127415 | 0.35 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr7_-_26603743 | 0.35 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr3_-_24681404 | 0.35 |
ENSDART00000161612
|
BX569789.1
|
|
| chr6_+_13045885 | 0.34 |
ENSDART00000104757
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr6_-_40768654 | 0.34 |
ENSDART00000184668
ENSDART00000146470 |
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr14_+_20893065 | 0.34 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr7_+_29512673 | 0.34 |
ENSDART00000173895
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr3_+_36424055 | 0.34 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr1_-_45157243 | 0.33 |
ENSDART00000131882
|
mucms1
|
mucin, multiple PTS and SEA group, member 1 |
| chr21_-_39024754 | 0.33 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr7_-_57637779 | 0.33 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr17_-_24890843 | 0.32 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr2_-_43135700 | 0.32 |
ENSDART00000098284
|
ftr14
|
finTRIM family, member 14 |
| chr11_+_14280598 | 0.32 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr8_+_19356072 | 0.32 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr9_+_3153717 | 0.32 |
ENSDART00000191958
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr17_+_25331576 | 0.32 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr5_+_12528693 | 0.31 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr24_+_20916751 | 0.31 |
ENSDART00000043193
|
cst14a.1
|
cystatin 14a, tandem duplicate 1 |
| chr11_-_40519886 | 0.31 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr19_-_340347 | 0.31 |
ENSDART00000139924
|
golph3l
|
golgi phosphoprotein 3-like |
| chr11_-_25853212 | 0.30 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr22_-_17652914 | 0.30 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr4_-_992063 | 0.30 |
ENSDART00000181630
ENSDART00000183898 ENSDART00000160902 |
naga
|
N-acetylgalactosaminidase, alpha |
| chr4_-_5826320 | 0.30 |
ENSDART00000165354
|
foxm1
|
forkhead box M1 |
| chr5_+_19867794 | 0.30 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr2_-_43151358 | 0.29 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr19_-_340641 | 0.29 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr19_-_32500373 | 0.29 |
ENSDART00000052104
|
fuca1.1
|
alpha-L-fucosidase 1, tandem duplicate 1 |
| chr20_-_26551210 | 0.29 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr13_+_228045 | 0.28 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr16_+_3982590 | 0.28 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr22_+_19528851 | 0.28 |
ENSDART00000145079
|
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr19_-_48391415 | 0.28 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr5_+_37087583 | 0.28 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr5_+_54400971 | 0.28 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr1_-_8652648 | 0.27 |
ENSDART00000138324
ENSDART00000141407 ENSDART00000054987 |
actb1
|
actin, beta 1 |
| chr8_+_15239549 | 0.27 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr15_-_29326254 | 0.27 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr1_-_47161996 | 0.27 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr8_-_52715911 | 0.27 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr5_+_27404946 | 0.27 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr8_-_17184482 | 0.26 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr11_+_14281100 | 0.26 |
ENSDART00000193664
ENSDART00000161597 |
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr16_+_38201840 | 0.26 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr7_-_73717082 | 0.26 |
ENSDART00000164301
ENSDART00000082625 |
BX664721.2
|
|
| chr21_+_5508167 | 0.26 |
ENSDART00000157702
|
ly6m4
|
lymphocyte antigen 6 family member M4 |
| chr6_-_33924883 | 0.26 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr17_+_45395846 | 0.26 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr22_+_3914318 | 0.26 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr17_-_11439815 | 0.26 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr9_-_42484444 | 0.25 |
ENSDART00000048320
ENSDART00000047653 |
tfpia
|
tissue factor pathway inhibitor a |
| chr4_-_13509946 | 0.25 |
ENSDART00000134720
|
ifng1-2
|
interferon, gamma 1-2 |
| chr24_-_10897511 | 0.25 |
ENSDART00000145593
ENSDART00000102484 ENSDART00000066784 |
fam49bb
|
family with sequence similarity 49, member Bb |
| chr24_+_41281265 | 0.25 |
ENSDART00000156920
ENSDART00000155151 |
nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr15_-_43284021 | 0.25 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr6_-_33925381 | 0.25 |
ENSDART00000137268
ENSDART00000145019 ENSDART00000141822 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr10_+_44677632 | 0.25 |
ENSDART00000171035
|
tpst2
|
tyrosylprotein sulfotransferase 2 |
| chr8_+_25342896 | 0.25 |
ENSDART00000129032
|
CR847543.1
|
|
| chr3_+_43826018 | 0.25 |
ENSDART00000166197
ENSDART00000165145 |
litaf
|
lipopolysaccharide-induced TNF factor |
| chr9_-_40011673 | 0.25 |
ENSDART00000184726
|
IKZF2
|
si:zfos-1425h8.1 |
| chr8_-_21103522 | 0.25 |
ENSDART00000100283
|
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr25_+_19238175 | 0.25 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr14_-_38946808 | 0.24 |
ENSDART00000139293
|
gla
|
galactosidase, alpha |
| chr19_-_19379084 | 0.24 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr20_-_54522660 | 0.24 |
ENSDART00000059872
|
ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr19_-_336777 | 0.24 |
ENSDART00000138837
|
golph3l
|
golgi phosphoprotein 3-like |
| chr16_-_6849754 | 0.24 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr19_-_15420678 | 0.24 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr11_-_21404044 | 0.24 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr5_+_38619813 | 0.24 |
ENSDART00000133314
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr1_+_58242498 | 0.24 |
ENSDART00000149091
|
ggt1l2.2
|
gamma-glutamyltransferase 1 like 2.2 |
| chr10_-_21953643 | 0.24 |
ENSDART00000188921
ENSDART00000193569 |
FO744833.2
|
|
| chr5_-_25084318 | 0.24 |
ENSDART00000089339
|
dph7
|
diphthamide biosynthesis 7 |
| chr8_-_39760427 | 0.23 |
ENSDART00000184336
ENSDART00000186500 |
si:ch211-170d8.8
|
si:ch211-170d8.8 |
| chr16_-_9802449 | 0.23 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr6_+_18298444 | 0.23 |
ENSDART00000166018
|
card14
|
caspase recruitment domain family, member 14 |
| chr3_+_59899452 | 0.23 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr6_-_38930726 | 0.23 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr14_+_35424539 | 0.23 |
ENSDART00000171809
ENSDART00000162185 |
sytl4
|
synaptotagmin-like 4 |
| chr20_+_48754045 | 0.23 |
ENSDART00000062578
|
zgc:110348
|
zgc:110348 |
| chr17_+_21126495 | 0.23 |
ENSDART00000022830
|
pygb
|
phosphorylase, glycogen; brain |
| chr7_+_55149001 | 0.23 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr8_-_23599096 | 0.22 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr25_+_5015019 | 0.22 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr25_+_37435335 | 0.22 |
ENSDART00000171602
|
chmp1a
|
charged multivesicular body protein 1A |
| chr19_+_40350468 | 0.22 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr7_-_19613388 | 0.22 |
ENSDART00000173664
|
si:ch211-212k18.13
|
si:ch211-212k18.13 |
| chr17_-_1705013 | 0.22 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr17_-_6508406 | 0.22 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr19_+_23296616 | 0.22 |
ENSDART00000134567
|
irgf1
|
immunity-related GTPase family, f1 |
| chr20_-_52902693 | 0.22 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr15_-_28904371 | 0.22 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr13_+_25200105 | 0.22 |
ENSDART00000039640
|
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr1_-_29658721 | 0.22 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr14_-_897874 | 0.22 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr5_+_28271412 | 0.22 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr25_-_16987020 | 0.22 |
ENSDART00000073419
|
ccnd2a
|
cyclin D2, a |
| chr15_-_23793641 | 0.21 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr2_-_43130812 | 0.21 |
ENSDART00000039079
ENSDART00000056162 |
ftr13
|
finTRIM family, member 13 |
| chr7_-_59054322 | 0.21 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr23_+_20669149 | 0.21 |
ENSDART00000138936
|
arhgef3l
|
Rho guanine nucleotide exchange factor (GEF) 3, like |
| chr1_-_8653385 | 0.21 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr21_-_40717678 | 0.21 |
ENSDART00000045124
|
pomp
|
proteasome maturation protein |
| chr5_+_38631687 | 0.21 |
ENSDART00000147280
|
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr5_+_1933131 | 0.21 |
ENSDART00000061693
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr13_-_22961605 | 0.21 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr3_+_19621034 | 0.21 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr23_+_27675581 | 0.21 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr2_-_27609189 | 0.20 |
ENSDART00000040555
|
lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr16_-_51888952 | 0.20 |
ENSDART00000186407
ENSDART00000175435 |
rpl28
|
ribosomal protein L28 |
| chr1_+_12335816 | 0.20 |
ENSDART00000067086
|
nansa
|
N-acetylneuraminic acid synthase a |
| chr15_+_41919484 | 0.20 |
ENSDART00000099821
ENSDART00000146246 |
nlrp16
|
NACHT, LRR and PYD domains-containing protein 16 |
| chr7_-_59564011 | 0.20 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr1_+_57787642 | 0.20 |
ENSDART00000127091
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr12_-_49151326 | 0.20 |
ENSDART00000153244
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr5_-_31470204 | 0.20 |
ENSDART00000130843
|
p2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr1_+_58375871 | 0.20 |
ENSDART00000166736
|
CU914631.1
|
|
| chr14_+_13453130 | 0.20 |
ENSDART00000054849
|
pls3
|
plastin 3 (T isoform) |
| chr25_-_12998202 | 0.20 |
ENSDART00000171661
|
ccl39.3
|
chemokine (C-C motif) ligand 39, duplicate 3 |
| chr7_-_56606752 | 0.20 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr10_+_10313024 | 0.20 |
ENSDART00000142895
ENSDART00000129952 |
urm1
|
ubiquitin related modifier 1 |
| chr25_+_3549401 | 0.19 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr10_+_7798087 | 0.19 |
ENSDART00000141824
|
sftpba
|
surfactant protein Ba |
| chr10_-_20650302 | 0.19 |
ENSDART00000142028
ENSDART00000064662 |
rassf2b
|
Ras association (RalGDS/AF-6) domain family member 2b |
| chr18_+_21071023 | 0.19 |
ENSDART00000100778
|
arpin
|
actin related protein 2/3 complex inhibitor |
| chr7_+_19850889 | 0.19 |
ENSDART00000100782
|
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr24_+_38534550 | 0.19 |
ENSDART00000105677
|
zgc:154125
|
zgc:154125 |
| chr23_+_16807209 | 0.19 |
ENSDART00000141966
|
zgc:114081
|
zgc:114081 |
| chr7_+_25053331 | 0.19 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr7_+_19762595 | 0.19 |
ENSDART00000130347
|
si:dkey-9k7.3
|
si:dkey-9k7.3 |
| chr10_+_7636811 | 0.19 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr15_-_41744509 | 0.19 |
ENSDART00000176952
ENSDART00000156690 |
ftr88
|
finTRIM family, member 88 |
| chr25_+_5068442 | 0.18 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr6_-_19270484 | 0.18 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr22_+_10163901 | 0.18 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr17_+_6585333 | 0.18 |
ENSDART00000124293
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr16_-_26855936 | 0.18 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr3_-_40528333 | 0.18 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr5_+_12743640 | 0.18 |
ENSDART00000081411
|
pole
|
polymerase (DNA directed), epsilon |
| chr21_+_21244046 | 0.18 |
ENSDART00000058306
ENSDART00000189665 ENSDART00000131890 |
lifrb
|
leukemia inhibitory factor receptor alpha b |
| chr5_+_19933356 | 0.18 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr17_-_9962578 | 0.18 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr20_+_46572550 | 0.18 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr8_+_26396552 | 0.18 |
ENSDART00000087151
|
amt
|
aminomethyltransferase |
| chr16_+_43368572 | 0.18 |
ENSDART00000032778
ENSDART00000193897 |
rnf144b
|
ring finger protein 144B |
| chr20_-_35706379 | 0.18 |
ENSDART00000152847
|
si:dkey-30j22.1
|
si:dkey-30j22.1 |
| chr20_-_23253630 | 0.18 |
ENSDART00000103365
|
ociad1
|
OCIA domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ehf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.2 | 0.5 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.8 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.3 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.1 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.2 | GO:0002320 | response to protozoan(GO:0001562) lymphoid progenitor cell differentiation(GO:0002320) defense response to protozoan(GO:0042832) |
| 0.1 | 0.2 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.2 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.4 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.9 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.1 | GO:0043102 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.4 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 1.0 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.5 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.2 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0060420 | box C/D snoRNP assembly(GO:0000492) regulation of heart growth(GO:0060420) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:0051125 | regulation of actin nucleation(GO:0051125) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.0 | 0.1 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.5 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.0 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 0.5 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.2 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.7 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.2 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.2 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 0.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.2 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.0 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |