Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
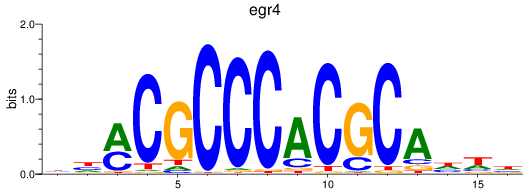
Results for egr4
Z-value: 0.83
Transcription factors associated with egr4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr4
|
ENSDARG00000077799 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr4 | dr11_v1_chr23_+_45579497_45579497 | -0.33 | 5.9e-01 | Click! |
Activity profile of egr4 motif
Sorted Z-values of egr4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_44963154 | 1.02 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr24_+_24831727 | 0.89 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr25_+_30298377 | 0.80 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr2_-_48753873 | 0.76 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr17_+_15297398 | 0.56 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr1_-_47431453 | 0.47 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr17_-_10025234 | 0.41 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr22_-_16180849 | 0.40 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr19_+_390298 | 0.32 |
ENSDART00000136361
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr2_+_35880600 | 0.31 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr3_+_16722014 | 0.27 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr23_+_16910071 | 0.27 |
ENSDART00000104793
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr4_-_9579299 | 0.26 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr25_-_207214 | 0.26 |
ENSDART00000193448
|
FP236318.3
|
|
| chr20_+_6659770 | 0.26 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr12_+_14079097 | 0.25 |
ENSDART00000078033
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr18_-_45617146 | 0.24 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr7_-_52498175 | 0.23 |
ENSDART00000129769
|
cgnl1
|
cingulin-like 1 |
| chr24_-_39354829 | 0.21 |
ENSDART00000169108
|
kcnj12a
|
potassium inwardly-rectifying channel, subfamily J, member 12a |
| chr8_+_8927870 | 0.20 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr22_-_6991514 | 0.20 |
ENSDART00000148233
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr5_-_43819663 | 0.19 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr3_-_5228137 | 0.19 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr16_-_40508624 | 0.18 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr10_-_15919839 | 0.18 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr5_-_2689753 | 0.16 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr18_+_8901846 | 0.16 |
ENSDART00000132109
ENSDART00000144247 |
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr8_+_45294767 | 0.16 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr19_+_342094 | 0.15 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr11_+_14049131 | 0.14 |
ENSDART00000172471
|
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr5_-_21030934 | 0.13 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr25_-_36827491 | 0.13 |
ENSDART00000170941
|
CU929365.1
|
|
| chr12_-_26537145 | 0.13 |
ENSDART00000138437
ENSDART00000163931 ENSDART00000132737 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr12_+_499881 | 0.12 |
ENSDART00000167527
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr6_+_2271559 | 0.12 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr11_+_12719944 | 0.12 |
ENSDART00000054837
ENSDART00000144954 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr8_+_22438398 | 0.12 |
ENSDART00000135145
|
zgc:153759
|
zgc:153759 |
| chr11_+_12720171 | 0.12 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr13_-_31397987 | 0.11 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr23_+_39606108 | 0.11 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr22_-_16275236 | 0.11 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr4_+_842010 | 0.10 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr2_+_16710889 | 0.09 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr10_+_19554604 | 0.08 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr5_+_42136359 | 0.07 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr22_+_6757741 | 0.07 |
ENSDART00000140815
|
CT583625.1
|
|
| chr21_-_41838284 | 0.07 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr3_+_23737795 | 0.07 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr20_+_27712714 | 0.07 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr23_+_9067131 | 0.06 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr4_+_70556298 | 0.06 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr14_+_15155684 | 0.06 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr16_-_12784373 | 0.06 |
ENSDART00000080396
|
foxj2
|
forkhead box J2 |
| chr6_-_23117348 | 0.06 |
ENSDART00000154941
ENSDART00000154252 |
ten1
|
TEN1 CST complex subunit |
| chr1_-_411331 | 0.06 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr16_+_19029297 | 0.05 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr21_+_33172526 | 0.05 |
ENSDART00000183532
|
arl3l1
|
ADP-ribosylation factor-like 3, like 1 |
| chr4_+_332709 | 0.05 |
ENSDART00000067486
|
tulp4a
|
tubby like protein 4a |
| chr13_-_49666615 | 0.05 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr6_+_40661703 | 0.04 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr21_-_43398457 | 0.04 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr22_+_1806235 | 0.03 |
ENSDART00000158966
|
znf1168
|
zinc finger protein 1168 |
| chr3_+_31897029 | 0.03 |
ENSDART00000146275
|
strada
|
ste20-related kinase adaptor alpha |
| chr21_-_43398122 | 0.03 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr17_+_48164536 | 0.03 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr4_+_28997595 | 0.03 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr14_-_38889840 | 0.03 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr1_-_10473630 | 0.02 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr9_-_48036 | 0.02 |
ENSDART00000165230
ENSDART00000171335 |
map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr22_-_5054773 | 0.02 |
ENSDART00000184207
|
matk
|
megakaryocyte-associated tyrosine kinase |
| chr14_+_40641350 | 0.02 |
ENSDART00000074506
|
tnmd
|
tenomodulin |
| chr3_+_22984098 | 0.02 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr23_+_804119 | 0.02 |
ENSDART00000148163
|
frmd4bb
|
FERM domain containing 4Bb |
| chr2_+_47906240 | 0.02 |
ENSDART00000122206
|
ftr23
|
finTRIM family, member 23 |
| chr5_-_49012569 | 0.02 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr11_+_3308656 | 0.02 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr21_+_6212844 | 0.02 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr15_+_36105581 | 0.01 |
ENSDART00000184099
|
col4a3
|
collagen, type IV, alpha 3 |
| chr11_-_277599 | 0.01 |
ENSDART00000187109
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr16_+_53519048 | 0.01 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr7_+_41340520 | 0.01 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr8_-_52859301 | 0.00 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr11_-_165288 | 0.00 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.3 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.6 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.5 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.3 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |