Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
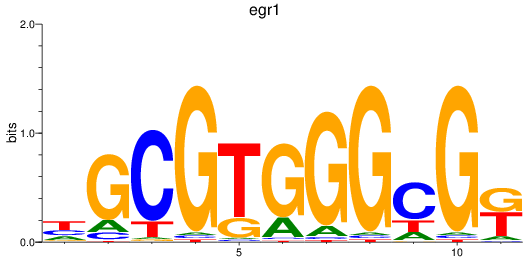
Results for egr1
Z-value: 0.81
Transcription factors associated with egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr1
|
ENSDARG00000037421 | early growth response 1 |
|
egr1
|
ENSDARG00000115563 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr1 | dr11_v1_chr14_-_21336385_21336385 | 0.29 | 6.3e-01 | Click! |
Activity profile of egr1 motif
Sorted Z-values of egr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_22535 | 1.74 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr20_-_54462551 | 0.96 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr2_-_48753873 | 0.82 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr23_+_37323962 | 0.67 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr2_+_47582681 | 0.65 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr2_+_47582488 | 0.63 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr24_+_24923166 | 0.61 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr2_+_38924975 | 0.59 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr25_+_34984333 | 0.53 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr9_-_18877597 | 0.51 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr18_+_18612388 | 0.50 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr23_+_41799748 | 0.50 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr17_-_14726824 | 0.45 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr13_+_12739283 | 0.42 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr20_-_45661049 | 0.42 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr3_-_61185746 | 0.40 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr1_-_7917062 | 0.38 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr13_-_21739142 | 0.38 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr6_-_35446110 | 0.37 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr8_+_24854600 | 0.37 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr23_+_20422661 | 0.36 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr21_+_28958471 | 0.36 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_-_20611039 | 0.36 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr16_-_6821927 | 0.35 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr23_+_2136344 | 0.35 |
ENSDART00000085290
ENSDART00000092757 |
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr12_+_47448558 | 0.34 |
ENSDART00000185689
ENSDART00000105328 |
fmn2b
|
formin 2b |
| chr9_-_54001502 | 0.34 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr10_-_32851847 | 0.34 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr1_+_32528097 | 0.33 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr20_-_915234 | 0.33 |
ENSDART00000164816
|
cnr1
|
cannabinoid receptor 1 |
| chr6_+_34512313 | 0.33 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr15_-_19250543 | 0.33 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr9_-_24413008 | 0.33 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr4_-_9586713 | 0.32 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr12_-_850457 | 0.32 |
ENSDART00000022688
|
tob1b
|
transducer of ERBB2, 1b |
| chr18_-_19103929 | 0.32 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr14_-_7888748 | 0.31 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr1_+_54673846 | 0.31 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr6_-_15604417 | 0.31 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr25_+_35706493 | 0.30 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr21_+_11415224 | 0.30 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr25_+_1304173 | 0.29 |
ENSDART00000155229
|
rxfp3.3b
|
relaxin/insulin-like family peptide receptor 3.3b |
| chr25_+_21177326 | 0.29 |
ENSDART00000149142
|
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr6_-_15604157 | 0.29 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr17_+_15983247 | 0.29 |
ENSDART00000154275
|
clmn
|
calmin |
| chr6_+_34511886 | 0.29 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr22_+_15979430 | 0.28 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr25_+_21176937 | 0.28 |
ENSDART00000163751
ENSDART00000188930 ENSDART00000187155 |
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr15_-_12319065 | 0.28 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr25_+_34888886 | 0.28 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr6_-_25181012 | 0.28 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr12_+_48511605 | 0.27 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr13_+_9432501 | 0.27 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr14_-_21218891 | 0.27 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr5_-_21524251 | 0.27 |
ENSDART00000176793
ENSDART00000040184 |
tenm1
|
teneurin transmembrane protein 1 |
| chr2_-_17827983 | 0.27 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr3_-_6767440 | 0.26 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr25_+_34889061 | 0.25 |
ENSDART00000136226
|
spire2
|
spire-type actin nucleation factor 2 |
| chr6_+_21740672 | 0.25 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr15_-_20939579 | 0.25 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr6_+_59808677 | 0.24 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr5_+_65492183 | 0.24 |
ENSDART00000162804
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr4_-_5019113 | 0.24 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr22_-_3564563 | 0.23 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr21_+_26620867 | 0.23 |
ENSDART00000176008
|
esrra
|
estrogen-related receptor alpha |
| chr15_+_3284684 | 0.23 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
| chr8_-_39903803 | 0.23 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr1_-_20271138 | 0.23 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr1_+_1599979 | 0.23 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr15_+_43906043 | 0.23 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr12_-_48943467 | 0.22 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr2_-_17828279 | 0.22 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr6_+_42819337 | 0.22 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr23_+_41800052 | 0.21 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr18_+_3169579 | 0.21 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr11_-_2594045 | 0.21 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr19_+_342094 | 0.21 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr23_-_31645760 | 0.20 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr24_+_32472155 | 0.20 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr15_-_14486534 | 0.20 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr2_-_155270 | 0.20 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr4_-_5018705 | 0.20 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr16_+_52105227 | 0.19 |
ENSDART00000150025
ENSDART00000097863 |
MAN1C1
|
si:ch73-373m9.1 |
| chr15_+_3284416 | 0.19 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr5_+_37032038 | 0.19 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr21_+_7900107 | 0.18 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr9_-_41401564 | 0.18 |
ENSDART00000059628
|
nab1b
|
NGFI-A binding protein 1b (EGR1 binding protein 1) |
| chr21_-_24865217 | 0.18 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr12_-_15620090 | 0.18 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr3_-_22829710 | 0.18 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr21_+_27513859 | 0.18 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr5_+_15667874 | 0.18 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr9_-_30363770 | 0.18 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr25_-_7999756 | 0.17 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr18_+_16246806 | 0.17 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr12_+_48851381 | 0.17 |
ENSDART00000187241
ENSDART00000187796 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr1_-_8917902 | 0.17 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr12_+_41510492 | 0.17 |
ENSDART00000170976
ENSDART00000176164 |
kif5bb
|
kinesin family member 5B, b |
| chr17_+_15983557 | 0.16 |
ENSDART00000190806
|
clmn
|
calmin |
| chr1_-_625875 | 0.16 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr25_+_12012 | 0.16 |
ENSDART00000170348
|
cntn1a
|
contactin 1a |
| chr2_+_50062165 | 0.16 |
ENSDART00000097939
ENSDART00000138214 |
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr20_+_32523576 | 0.16 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr14_+_7939398 | 0.16 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr2_+_57864307 | 0.15 |
ENSDART00000191981
|
plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr2_-_24603325 | 0.15 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr18_+_15106518 | 0.15 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr21_-_24865454 | 0.15 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr3_-_42016693 | 0.15 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr11_-_277599 | 0.14 |
ENSDART00000187109
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr11_-_37880492 | 0.14 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr6_+_11026866 | 0.14 |
ENSDART00000189619
|
abcc6a
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6a |
| chr2_-_48966431 | 0.14 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr25_+_29688671 | 0.14 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr23_+_44665230 | 0.14 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr8_-_14144707 | 0.14 |
ENSDART00000148061
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr25_-_37501371 | 0.14 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr8_+_28724692 | 0.14 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr20_+_27087539 | 0.13 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr20_-_54377933 | 0.13 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr23_-_27547931 | 0.13 |
ENSDART00000144419
|
larp4aa
|
La ribonucleoprotein domain family, member 4Aa |
| chr5_-_67241633 | 0.13 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr19_-_3303995 | 0.13 |
ENSDART00000105150
|
si:ch211-133n4.9
|
si:ch211-133n4.9 |
| chr14_-_4044545 | 0.13 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr22_+_10713713 | 0.12 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr20_-_54381034 | 0.12 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr6_+_11026354 | 0.12 |
ENSDART00000065433
|
abcc6a
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6a |
| chr2_+_23701613 | 0.12 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr1_-_10473630 | 0.12 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr7_+_31051213 | 0.12 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr24_+_3328354 | 0.12 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr25_-_6389713 | 0.12 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr22_-_34872533 | 0.12 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr13_-_49666615 | 0.11 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr4_-_9579299 | 0.11 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr8_+_20438884 | 0.11 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr1_-_29761086 | 0.10 |
ENSDART00000136760
|
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr9_+_38075851 | 0.10 |
ENSDART00000135314
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr13_-_479129 | 0.10 |
ENSDART00000159803
ENSDART00000082127 |
heatr5b
|
HEAT repeat containing 5B |
| chr7_+_31051603 | 0.10 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr17_-_25831569 | 0.10 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr1_+_12295142 | 0.10 |
ENSDART00000158595
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr3_+_19299309 | 0.10 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr20_-_26383368 | 0.10 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr13_+_30172142 | 0.10 |
ENSDART00000169917
ENSDART00000182091 |
pald1b
|
phosphatase domain containing, paladin 1b |
| chr7_+_20017211 | 0.09 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr1_+_55293424 | 0.09 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr11_+_77526 | 0.09 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr1_-_50438247 | 0.09 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr8_+_45294767 | 0.08 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr20_+_28803642 | 0.08 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr7_-_12596727 | 0.08 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr24_+_26379441 | 0.08 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr12_-_37449396 | 0.08 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr22_+_38192568 | 0.08 |
ENSDART00000126323
|
cp
|
ceruloplasmin |
| chr11_+_25296366 | 0.07 |
ENSDART00000065949
|
top1l
|
DNA topoisomerase I, like |
| chr1_-_48922273 | 0.07 |
ENSDART00000150254
|
si:ch211-112b1.2
|
si:ch211-112b1.2 |
| chr4_-_23839789 | 0.07 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr11_+_2699951 | 0.07 |
ENSDART00000082512
|
tmem167b
|
transmembrane protein 167B |
| chr16_+_4138331 | 0.07 |
ENSDART00000192403
ENSDART00000193036 |
mtf1
|
metal-regulatory transcription factor 1 |
| chr19_-_2115040 | 0.07 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr19_+_935565 | 0.07 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr23_+_9057999 | 0.06 |
ENSDART00000091899
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr14_+_36220479 | 0.06 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr23_+_45822935 | 0.06 |
ENSDART00000161892
|
vdra
|
vitamin D receptor a |
| chr1_-_17693273 | 0.06 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr9_+_45227028 | 0.06 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr3_+_45687266 | 0.06 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr3_+_16722014 | 0.05 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr11_+_2855430 | 0.05 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr18_-_50956280 | 0.05 |
ENSDART00000058457
|
st7
|
suppression of tumorigenicity 7 |
| chr2_+_20793982 | 0.05 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr13_+_25486608 | 0.05 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr1_-_59422880 | 0.05 |
ENSDART00000167244
|
si:ch211-188p14.2
|
si:ch211-188p14.2 |
| chr3_-_15444396 | 0.05 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr6_+_2271559 | 0.05 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr19_+_2619444 | 0.05 |
ENSDART00000169483
|
fam126a
|
family with sequence similarity 126, member A |
| chr18_-_50845804 | 0.05 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr19_-_48039400 | 0.05 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr3_-_37571601 | 0.04 |
ENSDART00000016407
|
arf2a
|
ADP-ribosylation factor 2a |
| chr9_+_51784922 | 0.04 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr2_+_23062085 | 0.04 |
ENSDART00000153745
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr10_+_23548674 | 0.04 |
ENSDART00000079686
|
zmp:0000001103
|
zmp:0000001103 |
| chr9_+_38369872 | 0.04 |
ENSDART00000193914
|
plcd4b
|
phospholipase C, delta 4b |
| chr14_+_12316581 | 0.04 |
ENSDART00000170115
ENSDART00000149757 |
si:ch211-125c23.3
cx31.7
|
si:ch211-125c23.3 connexin 31.7 |
| chr23_+_25856541 | 0.04 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr21_+_103194 | 0.04 |
ENSDART00000162755
|
zer1
|
zyg-11 related, cell cycle regulator |
| chr20_+_54356272 | 0.04 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr10_+_11282883 | 0.03 |
ENSDART00000135355
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr6_+_42818963 | 0.03 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr25_-_207214 | 0.03 |
ENSDART00000193448
|
FP236318.3
|
|
| chr19_+_43504480 | 0.03 |
ENSDART00000159421
|
BX649384.3
|
|
| chr20_-_48172556 | 0.03 |
ENSDART00000097888
|
CABZ01059099.1
|
|
| chr20_+_13894123 | 0.03 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr5_+_66170479 | 0.03 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr7_-_24995631 | 0.03 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr12_+_499881 | 0.03 |
ENSDART00000167527
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr22_+_34784075 | 0.03 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr1_+_49435017 | 0.03 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr10_-_14920989 | 0.03 |
ENSDART00000184617
|
smad2
|
SMAD family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.5 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.3 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.4 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.6 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:2000253 | cannabinoid signaling pathway(GO:0038171) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 1.1 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.4 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.7 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 1.1 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |