Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
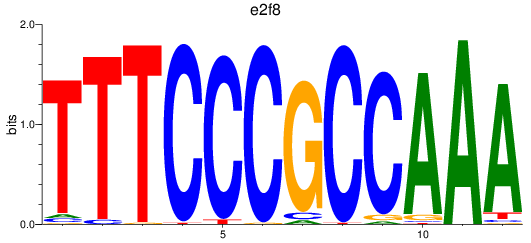
Results for e2f8
Z-value: 0.93
Transcription factors associated with e2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f8
|
ENSDARG00000057323 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f8 | dr11_v1_chr7_-_16597130_16597130 | 0.18 | 7.7e-01 | Click! |
Activity profile of e2f8 motif
Sorted Z-values of e2f8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_14155781 | 0.69 |
ENSDART00000169232
|
nr0b2b
|
nuclear receptor subfamily 0, group B, member 2b |
| chr14_+_46223458 | 0.60 |
ENSDART00000173428
|
cabp2b
|
calcium binding protein 2b |
| chr24_-_2829049 | 0.56 |
ENSDART00000164913
|
PPP1R3G
|
si:ch211-152c8.5 |
| chr4_-_2380173 | 0.55 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr7_+_24814866 | 0.54 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr9_+_33220342 | 0.50 |
ENSDART00000100893
ENSDART00000113451 |
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr11_-_29685853 | 0.45 |
ENSDART00000128468
|
nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr6_-_40071899 | 0.44 |
ENSDART00000034730
|
sec61a1
|
Sec61 translocon alpha 1 subunit |
| chr3_-_5016893 | 0.37 |
ENSDART00000165968
|
CSDC2 (1 of many)
|
cold shock domain containing C2 |
| chr6_+_50451337 | 0.36 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr16_-_4640539 | 0.36 |
ENSDART00000076955
ENSDART00000131949 |
cyp4t8
|
cytochrome P450, family 4, subfamily T, polypeptide 8 |
| chr2_+_5793908 | 0.35 |
ENSDART00000145219
|
slc35d1b
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1b |
| chr3_-_49514874 | 0.34 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr7_-_55648336 | 0.33 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr1_-_18811517 | 0.32 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr6_-_33916756 | 0.32 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr12_+_31616412 | 0.32 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr11_-_26576754 | 0.32 |
ENSDART00000191733
ENSDART00000002846 |
st3gal8
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 8 |
| chr2_+_32016256 | 0.30 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr7_+_69905307 | 0.30 |
ENSDART00000035907
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr13_+_46941930 | 0.29 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr21_-_11970199 | 0.27 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr20_-_38827623 | 0.27 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr17_-_36529016 | 0.27 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr2_+_32016516 | 0.27 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr3_+_25999477 | 0.26 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr19_+_32603627 | 0.26 |
ENSDART00000052086
|
tmem54b
|
transmembrane protein 54b |
| chr23_+_20803270 | 0.25 |
ENSDART00000097381
|
zgc:154075
|
zgc:154075 |
| chr5_-_37959874 | 0.23 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr3_-_35865040 | 0.23 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr20_+_13141408 | 0.23 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr17_-_36529449 | 0.22 |
ENSDART00000187252
|
colec11
|
collectin sub-family member 11 |
| chr14_-_28566238 | 0.21 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr9_-_2892045 | 0.21 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr18_-_34171280 | 0.21 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr3_+_43086548 | 0.21 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr4_-_7869731 | 0.20 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr6_-_18531760 | 0.20 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr23_+_44049509 | 0.19 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr18_-_34170918 | 0.18 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_+_38321039 | 0.18 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr19_-_44064842 | 0.18 |
ENSDART00000151541
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr10_+_6884123 | 0.17 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr15_-_36055401 | 0.17 |
ENSDART00000154476
|
col4a4
|
collagen, type IV, alpha 4 |
| chr24_-_35561672 | 0.17 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr7_+_71535045 | 0.17 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr9_-_2892250 | 0.16 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr13_+_48359573 | 0.15 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr13_+_2894536 | 0.15 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr8_+_52415603 | 0.15 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr11_-_4302251 | 0.15 |
ENSDART00000182554
|
CT033790.1
|
|
| chr1_+_59142783 | 0.15 |
ENSDART00000152669
|
gpr108
|
G protein-coupled receptor 108 |
| chr7_-_73845736 | 0.15 |
ENSDART00000193414
|
zgc:173552
|
zgc:173552 |
| chr14_-_32631519 | 0.15 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase phospholipid transporting 11C |
| chr7_+_26545911 | 0.14 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr13_-_47403154 | 0.14 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr6_+_21062416 | 0.14 |
ENSDART00000141643
|
si:dkey-91f15.1
|
si:dkey-91f15.1 |
| chr18_+_33788340 | 0.14 |
ENSDART00000136950
|
si:dkey-145c18.2
|
si:dkey-145c18.2 |
| chr23_+_42434348 | 0.14 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr25_-_35140746 | 0.13 |
ENSDART00000129969
|
si:ch211-113a14.19
|
si:ch211-113a14.19 |
| chr18_+_35842933 | 0.13 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr22_-_22301672 | 0.13 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr16_+_33953644 | 0.13 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr8_-_44238526 | 0.13 |
ENSDART00000061115
ENSDART00000132473 ENSDART00000138019 ENSDART00000133021 |
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr8_+_41038141 | 0.13 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr11_-_11792766 | 0.13 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr5_+_22459087 | 0.12 |
ENSDART00000134781
|
BX546499.1
|
|
| chr2_-_6104035 | 0.12 |
ENSDART00000053867
|
zte38
|
zebrafish testis-expressed 38 |
| chr25_-_35110695 | 0.12 |
ENSDART00000099859
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr7_+_5956937 | 0.12 |
ENSDART00000170763
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr10_+_10386435 | 0.12 |
ENSDART00000179214
ENSDART00000189799 ENSDART00000193875 |
sardh
|
sarcosine dehydrogenase |
| chr25_-_35145920 | 0.12 |
ENSDART00000157137
|
si:dkey-261m9.19
|
si:dkey-261m9.19 |
| chr16_+_53278406 | 0.12 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr1_-_46505310 | 0.12 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr7_-_73854476 | 0.12 |
ENSDART00000186481
|
zgc:173552
|
zgc:173552 |
| chr24_-_13349802 | 0.11 |
ENSDART00000164729
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr10_-_37487612 | 0.11 |
ENSDART00000167964
ENSDART00000171326 |
si:dkey-65j4.2
|
si:dkey-65j4.2 |
| chr25_+_25464630 | 0.11 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr20_+_2732992 | 0.11 |
ENSDART00000152697
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr3_-_14571514 | 0.11 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr22_-_38274188 | 0.11 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr13_+_8255106 | 0.11 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr25_+_35133404 | 0.11 |
ENSDART00000188505
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr19_-_2582858 | 0.11 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr7_+_26545502 | 0.11 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr10_+_26646104 | 0.11 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr4_+_74943111 | 0.11 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr23_+_39558508 | 0.11 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr5_+_67390645 | 0.11 |
ENSDART00000014822
|
ebf2
|
early B cell factor 2 |
| chr20_+_53368611 | 0.10 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr21_+_11244068 | 0.10 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr2_+_7818368 | 0.10 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr24_-_13349464 | 0.10 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr15_+_14202114 | 0.10 |
ENSDART00000169369
|
si:dkey-243k1.3
|
si:dkey-243k1.3 |
| chr1_-_34450622 | 0.10 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr7_-_6355459 | 0.10 |
ENSDART00000172898
|
CU457819.1
|
|
| chr25_-_35109536 | 0.10 |
ENSDART00000185229
|
CU302436.5
|
|
| chr9_-_48407408 | 0.10 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr19_+_42660158 | 0.10 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr4_-_74062634 | 0.10 |
ENSDART00000105992
|
si:cabz01069012.2
|
si:cabz01069012.2 |
| chr1_-_58505626 | 0.10 |
ENSDART00000171304
|
si:ch73-236c18.8
|
si:ch73-236c18.8 |
| chr12_+_18234557 | 0.10 |
ENSDART00000130741
|
fam20cb
|
family with sequence similarity 20, member Cb |
| chr7_-_6345507 | 0.10 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr7_+_29509255 | 0.10 |
ENSDART00000076172
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr23_-_29357764 | 0.10 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr8_-_12403077 | 0.09 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr4_-_16644708 | 0.09 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr17_+_2727807 | 0.09 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr19_-_30359059 | 0.09 |
ENSDART00000103471
|
khdrbs1b
|
KH domain containing, RNA binding, signal transduction associated 1b |
| chr9_-_49964810 | 0.09 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr7_-_64152087 | 0.09 |
ENSDART00000165234
|
CR354398.1
|
|
| chr4_-_74069252 | 0.09 |
ENSDART00000112802
|
si:cabz01069012.2
|
si:cabz01069012.2 |
| chr25_+_32525131 | 0.09 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr8_+_45186989 | 0.09 |
ENSDART00000143861
|
si:ch211-220m6.4
|
si:ch211-220m6.4 |
| chr7_-_804515 | 0.09 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr22_-_4439311 | 0.09 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr7_-_6362460 | 0.09 |
ENSDART00000129239
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr5_-_30382925 | 0.08 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr15_-_4596623 | 0.08 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr7_+_66565572 | 0.08 |
ENSDART00000180250
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr15_+_5628363 | 0.08 |
ENSDART00000148193
|
mrap
|
melanocortin 2 receptor accessory protein |
| chr22_+_8753365 | 0.08 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr2_-_57378748 | 0.08 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr10_+_573667 | 0.08 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr4_-_8014463 | 0.08 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr24_+_26432541 | 0.08 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr7_-_6415991 | 0.08 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr2_+_35728033 | 0.07 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr13_-_45942000 | 0.07 |
ENSDART00000158534
ENSDART00000171822 ENSDART00000166442 |
si:ch211-62a1.4
|
si:ch211-62a1.4 |
| chr15_-_17618800 | 0.07 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr20_+_18703454 | 0.07 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr4_-_7231480 | 0.07 |
ENSDART00000159436
|
si:ch211-170p16.1
|
si:ch211-170p16.1 |
| chr3_+_14571813 | 0.07 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr2_-_53500424 | 0.07 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr3_+_8826905 | 0.07 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr6_-_28345002 | 0.07 |
ENSDART00000158955
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr13_-_18011168 | 0.07 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr2_+_48074243 | 0.07 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr6_+_38896158 | 0.07 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr4_-_58978527 | 0.07 |
ENSDART00000150408
|
si:ch211-64i20.5_2
|
si:ch211-64i20.5_2 |
| chr4_-_73488406 | 0.07 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr22_+_18349794 | 0.06 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr22_-_6959769 | 0.06 |
ENSDART00000105866
|
si:ch73-213k20.5
|
si:ch73-213k20.5 |
| chr3_-_53092509 | 0.06 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr18_+_19972853 | 0.06 |
ENSDART00000180071
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr11_+_45092866 | 0.06 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr25_-_35102781 | 0.06 |
ENSDART00000180881
ENSDART00000153747 |
si:dkey-108k21.24
|
si:dkey-108k21.24 |
| chr8_-_44363596 | 0.06 |
ENSDART00000123658
|
si:busm1-228j01.4
|
si:busm1-228j01.4 |
| chr7_+_34453185 | 0.06 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr23_+_29358188 | 0.06 |
ENSDART00000189242
|
tardbpl
|
TAR DNA binding protein, like |
| chr6_-_43817761 | 0.06 |
ENSDART00000183945
|
foxp1b
|
forkhead box P1b |
| chr12_-_30345444 | 0.06 |
ENSDART00000152985
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr2_+_17055069 | 0.06 |
ENSDART00000115078
|
thpo
|
thrombopoietin |
| chr7_+_41812190 | 0.05 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr15_-_44637492 | 0.05 |
ENSDART00000170936
|
alp3
|
alkaline phosphatase 3 |
| chr18_+_27571448 | 0.05 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr13_+_48358467 | 0.05 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr1_+_6243906 | 0.05 |
ENSDART00000127329
|
gtf3c3
|
general transcription factor IIIC, polypeptide 3 |
| chr7_+_66565930 | 0.05 |
ENSDART00000154597
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr1_+_38858399 | 0.05 |
ENSDART00000165454
|
CU915762.1
|
|
| chr7_+_41812817 | 0.05 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr24_-_30843250 | 0.05 |
ENSDART00000162920
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr4_-_64123545 | 0.05 |
ENSDART00000170040
|
BX914205.2
|
|
| chr22_+_25590391 | 0.05 |
ENSDART00000178133
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr8_+_8196087 | 0.04 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr23_-_16981899 | 0.04 |
ENSDART00000125472
ENSDART00000142297 ENSDART00000143180 |
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr11_+_24290130 | 0.04 |
ENSDART00000029695
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr16_+_29558530 | 0.04 |
ENSDART00000088146
|
ensab
|
endosulfine alpha b |
| chr21_-_2360906 | 0.04 |
ENSDART00000171645
|
si:ch73-299h12.9
|
si:ch73-299h12.9 |
| chr15_-_17619306 | 0.04 |
ENSDART00000184011
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr11_-_18107447 | 0.04 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr19_+_40069524 | 0.04 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr7_+_41812636 | 0.04 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr11_-_18017918 | 0.03 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr7_-_23971497 | 0.03 |
ENSDART00000173603
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr22_+_8753092 | 0.03 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr19_-_32042105 | 0.03 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr19_-_340641 | 0.03 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr7_+_1550966 | 0.03 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr2_+_9755422 | 0.03 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr15_+_618081 | 0.03 |
ENSDART00000181518
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr3_+_5504990 | 0.03 |
ENSDART00000143626
|
si:ch73-264i18.2
|
si:ch73-264i18.2 |
| chr23_+_10805188 | 0.03 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr2_+_37424261 | 0.03 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr23_+_29357790 | 0.03 |
ENSDART00000139223
ENSDART00000141928 |
tardbpl
|
TAR DNA binding protein, like |
| chr1_-_28890028 | 0.03 |
ENSDART00000075500
|
poglut1
|
protein O-glucosyltransferase 1 |
| chr16_-_9453591 | 0.02 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr2_-_54946834 | 0.02 |
ENSDART00000192280
|
CABZ01086574.1
|
|
| chr2_-_20599315 | 0.02 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr2_-_45027754 | 0.02 |
ENSDART00000168469
|
vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr23_-_30045661 | 0.02 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr6_-_8466717 | 0.02 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr7_+_57725708 | 0.02 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr1_+_9144958 | 0.02 |
ENSDART00000046507
|
rasd3
|
RASD family member 3 |
| chr19_-_340347 | 0.02 |
ENSDART00000139924
|
golph3l
|
golgi phosphoprotein 3-like |
| chr7_-_39360325 | 0.02 |
ENSDART00000098033
ENSDART00000173695 ENSDART00000173466 ENSDART00000173734 |
ambra1a
|
autophagy/beclin-1 regulator 1a |
| chr1_+_27052530 | 0.02 |
ENSDART00000184065
|
bnc2
|
basonuclin 2 |
| chr5_-_19394440 | 0.02 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr23_+_21482571 | 0.02 |
ENSDART00000187368
|
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr25_-_17579701 | 0.01 |
ENSDART00000073684
|
mmp15a
|
matrix metallopeptidase 15a |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.5 | GO:0008584 | male gonad development(GO:0008584) |
| 0.1 | 0.3 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.3 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.5 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.4 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.7 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.2 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.0 | 0.1 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0009217 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.4 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0008406 | gonad development(GO:0008406) |
| 0.0 | 0.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.4 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.2 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |