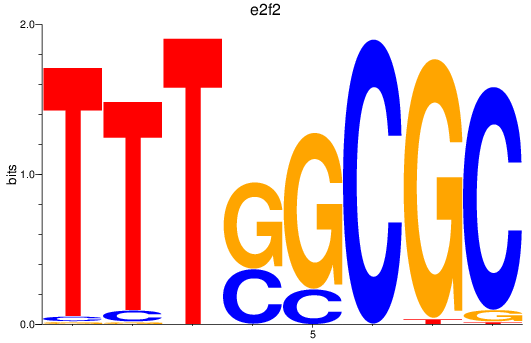
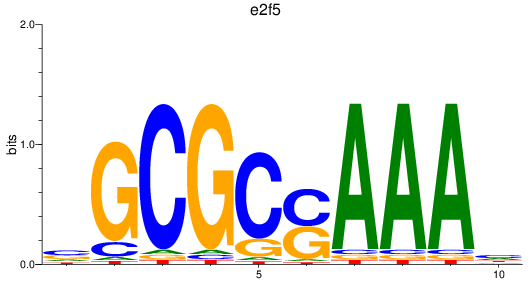
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for e2f2_e2f5
Z-value: 2.65
Transcription factors associated with e2f2_e2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f2
|
ENSDARG00000079233 | si_ch211-160f23.5 |
|
e2f5
|
ENSDARG00000038812 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F2 | dr11_v1_chr17_-_27266053_27266053 | 0.52 | 3.7e-01 | Click! |
| e2f5 | dr11_v1_chr2_-_31686353_31686403 | -0.02 | 9.8e-01 | Click! |
Activity profile of e2f2_e2f5 motif
Sorted Z-values of e2f2_e2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_904850 | 4.28 |
ENSDART00000161847
|
si:ch73-208h1.1
|
si:ch73-208h1.1 |
| chr9_-_2892250 | 3.14 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr23_+_31815423 | 1.92 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr20_+_39344889 | 1.90 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr3_+_25999477 | 1.74 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr9_-_2892045 | 1.73 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr24_-_35561672 | 1.61 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr11_-_11792766 | 1.59 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr20_+_34770197 | 1.55 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr1_+_19538299 | 1.47 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr3_-_54607166 | 1.44 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr13_+_8255106 | 1.31 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr8_+_52415603 | 1.29 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr8_-_1219815 | 1.21 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr19_-_47571797 | 1.19 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_-_26490186 | 1.17 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr7_+_71535045 | 1.01 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr20_+_23440632 | 0.99 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr10_-_21362071 | 0.99 |
ENSDART00000125167
|
avd
|
avidin |
| chr6_+_12968101 | 0.98 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr18_-_5595546 | 0.97 |
ENSDART00000191825
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr8_-_4760723 | 0.96 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr19_-_47571456 | 0.95 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr20_-_23439011 | 0.94 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr15_-_17099560 | 0.91 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr25_-_35095129 | 0.90 |
ENSDART00000099866
ENSDART00000099868 |
kif15
|
kinesin family member 15 |
| chr8_-_12403077 | 0.90 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr6_+_32046202 | 0.89 |
ENSDART00000156552
|
si:dkey-148h10.5
|
si:dkey-148h10.5 |
| chr3_+_37112693 | 0.88 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr22_-_4439311 | 0.85 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr21_+_22558187 | 0.84 |
ENSDART00000167599
|
chek1
|
checkpoint kinase 1 |
| chr4_-_77561679 | 0.81 |
ENSDART00000180809
|
AL935186.9
|
|
| chr6_+_59029485 | 0.80 |
ENSDART00000050140
|
CABZ01088367.1
|
|
| chr17_+_25481466 | 0.78 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr24_+_11908480 | 0.76 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr19_-_2822372 | 0.76 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr13_-_13030851 | 0.76 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr11_-_309420 | 0.76 |
ENSDART00000173185
|
poc1a
|
POC1 centriolar protein A |
| chr20_-_29499363 | 0.74 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr4_+_76748500 | 0.74 |
ENSDART00000075607
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr22_-_3152357 | 0.74 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr3_+_22335030 | 0.73 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr6_+_50451337 | 0.73 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr24_+_11908833 | 0.72 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr5_-_19014589 | 0.71 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr21_-_233282 | 0.71 |
ENSDART00000157684
|
bxdc2
|
brix domain containing 2 |
| chr7_+_55518519 | 0.70 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr16_-_43335914 | 0.70 |
ENSDART00000111963
|
atad2
|
ATPase family, AAA domain containing 2 |
| chr5_+_12743640 | 0.68 |
ENSDART00000081411
|
pole
|
polymerase (DNA directed), epsilon |
| chr20_-_23440955 | 0.68 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr20_-_29498178 | 0.68 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr17_+_8799661 | 0.67 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr7_+_34236238 | 0.66 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr15_-_2954443 | 0.66 |
ENSDART00000161053
|
zgc:153184
|
zgc:153184 |
| chr6_+_33931740 | 0.66 |
ENSDART00000130492
ENSDART00000151213 |
orc1
|
origin recognition complex, subunit 1 |
| chr24_+_23840821 | 0.64 |
ENSDART00000128595
ENSDART00000127188 |
pola1
|
polymerase (DNA directed), alpha 1 |
| chr20_+_4392687 | 0.64 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr3_+_26244353 | 0.63 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr6_-_1762191 | 0.61 |
ENSDART00000167928
|
orc4
|
origin recognition complex, subunit 4 |
| chr19_-_7144548 | 0.56 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr2_-_44777592 | 0.55 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr7_-_2143313 | 0.54 |
ENSDART00000173871
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr25_-_13403726 | 0.54 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr25_+_186583 | 0.53 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr1_+_58375871 | 0.53 |
ENSDART00000166736
|
CU914631.1
|
|
| chr1_-_59216197 | 0.52 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr2_-_44344321 | 0.52 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr1_+_9199031 | 0.52 |
ENSDART00000092058
ENSDART00000182771 |
chtf18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr1_-_8553165 | 0.51 |
ENSDART00000135197
ENSDART00000054981 |
zgc:112980
|
zgc:112980 |
| chr7_-_6464225 | 0.51 |
ENSDART00000130760
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr13_-_36863018 | 0.50 |
ENSDART00000023449
ENSDART00000149011 |
pygl
|
phosphorylase, glycogen, liver |
| chr13_-_856525 | 0.48 |
ENSDART00000143356
|
TMEM14A
|
zgc:163080 |
| chr19_-_29294457 | 0.47 |
ENSDART00000130815
ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr5_+_32345187 | 0.46 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr13_-_35908275 | 0.45 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr2_-_29923403 | 0.44 |
ENSDART00000144672
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr2_+_44512324 | 0.44 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr17_+_8799451 | 0.43 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr4_-_5831522 | 0.43 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr1_-_46663997 | 0.42 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr8_-_13972626 | 0.40 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr7_+_71586485 | 0.39 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr10_+_29849497 | 0.39 |
ENSDART00000099994
ENSDART00000132212 |
hspa8
|
heat shock protein 8 |
| chr3_-_16110351 | 0.39 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr13_+_24584401 | 0.39 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr16_-_9802449 | 0.39 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr16_-_14587332 | 0.39 |
ENSDART00000012479
|
dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr22_+_1708760 | 0.38 |
ENSDART00000170471
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr3_-_26244256 | 0.38 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr9_-_30555725 | 0.38 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr7_+_1550966 | 0.38 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_-_2153147 | 0.38 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr16_+_33143503 | 0.37 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr17_+_4325693 | 0.37 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr5_+_69868911 | 0.37 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr10_-_35153388 | 0.37 |
ENSDART00000188132
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr19_+_48060464 | 0.36 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr24_-_11908115 | 0.36 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr18_-_30499489 | 0.36 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr22_-_10752471 | 0.35 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr4_+_77933084 | 0.35 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr21_-_25741096 | 0.34 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr6_-_51573975 | 0.34 |
ENSDART00000073865
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr4_+_18441988 | 0.34 |
ENSDART00000040827
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr7_+_41812636 | 0.34 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr22_-_10591876 | 0.33 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr6_+_7533601 | 0.33 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr21_-_27413294 | 0.33 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr13_-_856701 | 0.32 |
ENSDART00000140423
|
TMEM14A
|
zgc:163080 |
| chr25_-_35140746 | 0.32 |
ENSDART00000129969
|
si:ch211-113a14.19
|
si:ch211-113a14.19 |
| chr14_-_49859747 | 0.32 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr12_-_3962372 | 0.31 |
ENSDART00000016791
|
eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr1_-_201004 | 0.31 |
ENSDART00000188956
|
FO681286.1
|
|
| chr13_-_14929236 | 0.31 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr10_+_38775408 | 0.31 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr7_+_19851422 | 0.31 |
ENSDART00000142970
ENSDART00000190027 |
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr3_-_16110100 | 0.30 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr7_+_24814866 | 0.30 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr19_-_12322356 | 0.30 |
ENSDART00000016128
|
ncaldb
|
neurocalcin delta b |
| chr16_+_50953547 | 0.30 |
ENSDART00000157526
|
si:dkeyp-97a10.1
|
si:dkeyp-97a10.1 |
| chr22_-_5822147 | 0.29 |
ENSDART00000011076
|
cers5
|
ceramide synthase 5 |
| chr10_+_38775959 | 0.29 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr19_-_2582858 | 0.29 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr8_+_30112655 | 0.29 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr5_-_22969424 | 0.29 |
ENSDART00000143869
ENSDART00000172549 |
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr20_-_54245256 | 0.29 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr2_+_48074243 | 0.29 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr20_+_263056 | 0.29 |
ENSDART00000132669
|
tube1
|
tubulin, epsilon 1 |
| chr7_+_41812190 | 0.29 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr16_+_68069 | 0.29 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr5_+_8256574 | 0.29 |
ENSDART00000171343
|
FP102192.1
|
|
| chr5_-_33255759 | 0.28 |
ENSDART00000085531
|
prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr25_+_36333993 | 0.28 |
ENSDART00000184379
|
CR354435.2
|
|
| chr13_-_35907768 | 0.28 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr17_-_27266053 | 0.28 |
ENSDART00000110903
|
E2F2
|
si:ch211-160f23.5 |
| chr2_-_29923630 | 0.28 |
ENSDART00000158844
ENSDART00000031130 |
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr16_+_40575742 | 0.27 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr8_-_38406831 | 0.27 |
ENSDART00000112991
ENSDART00000191445 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr19_+_43604643 | 0.27 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr5_+_27488975 | 0.27 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr3_-_35865040 | 0.27 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr2_-_38363017 | 0.27 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr5_+_22969651 | 0.27 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr25_-_12824656 | 0.27 |
ENSDART00000171801
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr8_+_20415824 | 0.27 |
ENSDART00000009081
ENSDART00000145444 |
mob3a
|
MOB kinase activator 3A |
| chr23_+_17839187 | 0.27 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr14_+_80685 | 0.26 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr14_+_94603 | 0.26 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr21_+_45386033 | 0.26 |
ENSDART00000151773
|
jade2
|
jade family PHD finger 2 |
| chr17_+_43889371 | 0.26 |
ENSDART00000156871
ENSDART00000154702 |
msh4
|
mutS homolog 4 |
| chr24_+_17345521 | 0.25 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr13_-_7432715 | 0.25 |
ENSDART00000158685
|
si:dkey-45k15.1
|
si:dkey-45k15.1 |
| chr3_+_29458517 | 0.25 |
ENSDART00000134258
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr19_+_9455218 | 0.24 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr9_-_12652984 | 0.24 |
ENSDART00000052256
|
sumo3b
|
small ubiquitin-like modifier 3b |
| chr4_-_9909371 | 0.24 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr3_+_22984098 | 0.24 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr11_+_18216404 | 0.23 |
ENSDART00000086437
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr10_+_21434649 | 0.23 |
ENSDART00000193938
ENSDART00000064558 |
etf1b
|
eukaryotic translation termination factor 1b |
| chr25_+_3326885 | 0.23 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr1_-_12126535 | 0.23 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr7_+_5913030 | 0.23 |
ENSDART00000160575
|
CU459186.1
|
Histone H3.2 |
| chr20_-_38827623 | 0.23 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr25_-_35144620 | 0.23 |
ENSDART00000185669
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr20_-_30376433 | 0.22 |
ENSDART00000190737
|
rps7
|
ribosomal protein S7 |
| chr16_+_41015781 | 0.22 |
ENSDART00000124543
|
dek
|
DEK proto-oncogene |
| chr10_+_14499201 | 0.22 |
ENSDART00000064901
|
katnal2
|
katanin p60 subunit A-like 2 |
| chr16_-_35586401 | 0.22 |
ENSDART00000169868
ENSDART00000166606 ENSDART00000162562 |
scmh1
|
Scm polycomb group protein homolog 1 |
| chr3_-_45777226 | 0.22 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr4_-_2545310 | 0.22 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr5_-_54395488 | 0.22 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr2_+_48073972 | 0.22 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr17_-_49407091 | 0.22 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr18_+_41560822 | 0.22 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr1_+_26677406 | 0.22 |
ENSDART00000183427
ENSDART00000180366 ENSDART00000181997 |
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr21_-_23017478 | 0.21 |
ENSDART00000024309
|
rb1
|
retinoblastoma 1 |
| chr1_+_50976975 | 0.21 |
ENSDART00000022290
ENSDART00000140982 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr20_-_29633507 | 0.21 |
ENSDART00000040292
|
cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr15_-_19334448 | 0.21 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| chr22_+_1930589 | 0.21 |
ENSDART00000159807
|
znf1153
|
zinc finger protein 1153 |
| chr14_+_46223458 | 0.21 |
ENSDART00000173428
|
cabp2b
|
calcium binding protein 2b |
| chr21_+_26522571 | 0.21 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr25_+_36327034 | 0.21 |
ENSDART00000073452
|
zgc:110216
|
zgc:110216 |
| chr7_-_37895771 | 0.20 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr2_-_47957673 | 0.20 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr21_-_30408775 | 0.20 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr25_-_35145920 | 0.20 |
ENSDART00000157137
|
si:dkey-261m9.19
|
si:dkey-261m9.19 |
| chr1_+_26676758 | 0.20 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr25_-_32363341 | 0.20 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr15_-_36347858 | 0.20 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr14_+_52563794 | 0.19 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr25_-_35047838 | 0.19 |
ENSDART00000155060
|
CU302436.3
|
|
| chr17_-_22573311 | 0.19 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr10_-_15644904 | 0.19 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr6_-_32045951 | 0.19 |
ENSDART00000016629
ENSDART00000139055 |
efcab7
|
EF-hand calcium binding domain 7 |
| chr3_-_32275975 | 0.19 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr19_-_20777351 | 0.19 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr17_+_26828027 | 0.19 |
ENSDART00000042060
|
jade1
|
jade family PHD finger 1 |
| chr4_+_70556298 | 0.18 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr7_-_69185124 | 0.18 |
ENSDART00000182217
ENSDART00000191359 |
usp10
|
ubiquitin specific peptidase 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f2_e2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.6 | 4.3 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.5 | 6.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.4 | 4.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 3.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.3 | 1.9 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.3 | 0.9 | GO:1905132 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.3 | 2.0 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.3 | 2.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 0.8 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 0.7 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 1.4 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.2 | 0.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.4 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.2 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.9 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.7 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 0.4 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 5.0 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 1.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.1 | GO:2000104 | negative regulation of DNA-dependent DNA replication(GO:2000104) |
| 0.1 | 0.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 1.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.4 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.2 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.4 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.5 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.6 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.5 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.1 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.0 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 1.1 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.2 | GO:0030516 | regulation of axon extension(GO:0030516) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.4 | 7.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 0.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 0.5 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 7.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 11.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 1.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 1.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.5 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 1.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.5 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.2 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 2.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.8 | GO:0016407 | acetyltransferase activity(GO:0016407) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 11.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 8.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.0 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 2.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME HIV LIFE CYCLE | Genes involved in HIV Life Cycle |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |