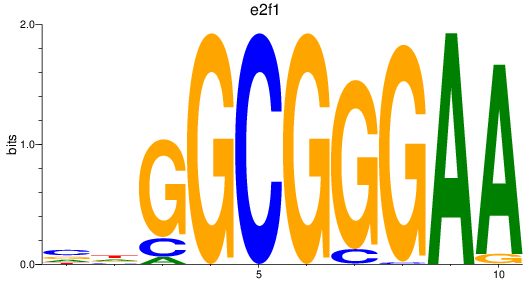
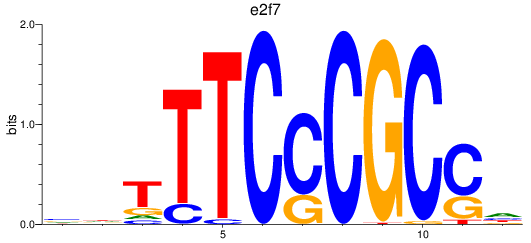
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for e2f1_e2f7
Z-value: 2.24
Transcription factors associated with e2f1_e2f7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f1
|
ENSDARG00000103868 | E2F transcription factor 1 |
|
e2f7
|
ENSDARG00000008986 | E2F transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f1 | dr11_v1_chr23_-_43486714_43486714 | 0.96 | 1.1e-02 | Click! |
| e2f7 | dr11_v1_chr4_-_2545310_2545345 | 0.29 | 6.4e-01 | Click! |
Activity profile of e2f1_e2f7 motif
Sorted Z-values of e2f1_e2f7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48391415 | 2.82 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr16_-_41990421 | 1.93 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr9_-_33081978 | 1.50 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr5_+_22970617 | 1.48 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr8_-_1219815 | 1.33 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr6_+_12968101 | 1.33 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr20_-_29498178 | 1.32 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_+_25999477 | 1.27 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr14_+_14841685 | 1.26 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr20_+_34770197 | 1.25 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr19_-_47570672 | 1.23 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_-_49514874 | 1.18 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr19_-_5358443 | 1.18 |
ENSDART00000105036
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr24_-_35561672 | 1.10 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr8_+_16407884 | 1.04 |
ENSDART00000133742
|
cdkn2c
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr7_+_24881680 | 1.02 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr12_+_46883785 | 1.01 |
ENSDART00000008312
|
fam53b
|
family with sequence similarity 53, member B |
| chr8_+_52415603 | 1.01 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr20_+_53521648 | 0.96 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr1_-_38170997 | 0.96 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr6_-_425378 | 0.96 |
ENSDART00000192190
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr13_-_86847 | 0.96 |
ENSDART00000158062
|
pole2
|
polymerase (DNA directed), epsilon 2 |
| chr15_-_2640966 | 0.95 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr8_+_16408385 | 0.92 |
ENSDART00000177231
|
cdkn2c
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr22_-_22301672 | 0.89 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr5_-_6567464 | 0.88 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr2_+_48074243 | 0.85 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr17_-_51195651 | 0.82 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr16_-_4769877 | 0.78 |
ENSDART00000149421
ENSDART00000054078 |
rpa2
|
replication protein A2 |
| chr6_-_33916756 | 0.77 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr15_-_1485086 | 0.76 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr17_-_51199558 | 0.75 |
ENSDART00000154532
|
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr10_+_24627683 | 0.73 |
ENSDART00000112652
|
slc46a3
|
solute carrier family 46, member 3 |
| chr20_+_53522059 | 0.72 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr23_+_17839187 | 0.71 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr20_-_54869006 | 0.71 |
ENSDART00000184817
|
CABZ01037174.1
|
|
| chr7_+_71586485 | 0.70 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr23_+_45025909 | 0.70 |
ENSDART00000188105
|
hmgb2b
|
high mobility group box 2b |
| chr13_-_35908275 | 0.69 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr16_-_4770233 | 0.69 |
ENSDART00000193228
|
rpa2
|
replication protein A2 |
| chr4_-_2380173 | 0.68 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr11_+_1551603 | 0.66 |
ENSDART00000185383
ENSDART00000121489 ENSDART00000040577 |
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr21_-_44731865 | 0.66 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr16_+_53252951 | 0.65 |
ENSDART00000126543
|
si:ch211-269k10.5
|
si:ch211-269k10.5 |
| chr14_-_22015232 | 0.64 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr2_+_48073972 | 0.63 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr7_-_16582187 | 0.63 |
ENSDART00000131726
|
e2f8
|
E2F transcription factor 8 |
| chr8_-_12403077 | 0.62 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr23_+_44049509 | 0.61 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr21_-_25213616 | 0.61 |
ENSDART00000122513
|
rfc2
|
replication factor C (activator 1) 2 |
| chr15_+_43333121 | 0.60 |
ENSDART00000161011
|
zgc:85932
|
zgc:85932 |
| chr1_-_7603734 | 0.60 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr25_+_32390794 | 0.60 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr8_+_54013199 | 0.59 |
ENSDART00000158497
|
CABZ01079663.1
|
|
| chr24_+_26328787 | 0.59 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr19_-_2582858 | 0.59 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr3_+_24537023 | 0.59 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr18_-_30499489 | 0.57 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr1_-_59216197 | 0.56 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr5_-_30535327 | 0.55 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr13_+_8255106 | 0.55 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr21_-_5393125 | 0.55 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr21_+_30721733 | 0.55 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr5_-_67365750 | 0.55 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr9_-_2892250 | 0.54 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr24_+_26329018 | 0.54 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr3_+_13332870 | 0.53 |
ENSDART00000113878
|
CU138547.1
|
|
| chr25_+_32496877 | 0.53 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr10_+_28428222 | 0.52 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr4_+_68776476 | 0.51 |
ENSDART00000169135
|
si:dkey-264f17.3
|
si:dkey-264f17.3 |
| chr18_+_30421528 | 0.51 |
ENSDART00000140908
|
gse1
|
Gse1 coiled-coil protein |
| chr11_+_45153104 | 0.51 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr25_-_35124576 | 0.50 |
ENSDART00000128017
|
zgc:171759
|
zgc:171759 |
| chr3_+_24571163 | 0.49 |
ENSDART00000123017
|
sp100.3
|
SP110 nuclear body protein, tandem duplicate 3 |
| chr6_-_6976096 | 0.49 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr17_-_20236228 | 0.49 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr23_-_36306337 | 0.48 |
ENSDART00000142760
ENSDART00000136929 ENSDART00000143340 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr19_-_8748571 | 0.47 |
ENSDART00000031173
|
rps27.1
|
ribosomal protein S27, isoform 1 |
| chr17_-_51199219 | 0.46 |
ENSDART00000154403
|
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr7_-_26306546 | 0.45 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr7_+_24814866 | 0.45 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr21_-_23017478 | 0.45 |
ENSDART00000024309
|
rb1
|
retinoblastoma 1 |
| chr11_-_45434959 | 0.45 |
ENSDART00000173106
ENSDART00000172767 ENSDART00000172933 ENSDART00000172986 |
rfc4
|
replication factor C (activator 1) 4 |
| chr15_-_1484795 | 0.45 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr18_-_8380090 | 0.44 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr2_-_59285085 | 0.44 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr10_-_15644904 | 0.44 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr2_-_122154 | 0.43 |
ENSDART00000156248
ENSDART00000004071 |
znfl2a
|
zinc finger-like gene 2a |
| chr8_-_2504714 | 0.43 |
ENSDART00000171559
|
rpl6
|
ribosomal protein L6 |
| chr11_-_3308569 | 0.42 |
ENSDART00000036581
|
cdk2
|
cyclin-dependent kinase 2 |
| chr23_-_3758637 | 0.42 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr13_+_48358467 | 0.42 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr7_+_1550966 | 0.42 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr13_+_41022502 | 0.41 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr5_-_67365333 | 0.41 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr16_-_12787029 | 0.41 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr13_+_42602406 | 0.41 |
ENSDART00000133388
ENSDART00000147996 |
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr6_+_112579 | 0.40 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr10_+_6010570 | 0.40 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr6_-_1762191 | 0.40 |
ENSDART00000167928
|
orc4
|
origin recognition complex, subunit 4 |
| chr23_-_36305874 | 0.39 |
ENSDART00000147598
ENSDART00000146986 ENSDART00000086985 ENSDART00000133259 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr18_+_17020967 | 0.39 |
ENSDART00000189168
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr18_+_20226843 | 0.39 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr24_+_10413484 | 0.39 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr9_-_30555725 | 0.39 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr19_+_25465025 | 0.38 |
ENSDART00000018553
|
rpa3
|
replication protein A3 |
| chr22_-_5655680 | 0.37 |
ENSDART00000159629
|
mcm2
|
minichromosome maintenance complex component 2 |
| chr2_-_59303338 | 0.37 |
ENSDART00000100987
ENSDART00000140840 |
ftr35
|
finTRIM family, member 35 |
| chr13_-_35907768 | 0.37 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr3_-_12227359 | 0.37 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr13_+_28785814 | 0.36 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr13_-_14929236 | 0.36 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr21_-_27413294 | 0.36 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr14_-_16775158 | 0.36 |
ENSDART00000113711
ENSDART00000144781 ENSDART00000160411 |
mrnip
|
MRN complex interacting protein |
| chr5_-_67365006 | 0.34 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr22_-_22301929 | 0.34 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr3_+_34988670 | 0.33 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr1_+_27690 | 0.33 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr12_-_4206869 | 0.33 |
ENSDART00000106572
|
si:dkey-32n7.9
|
si:dkey-32n7.9 |
| chr23_+_36306539 | 0.32 |
ENSDART00000053267
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr11_+_3308656 | 0.32 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr25_+_35051656 | 0.32 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr25_+_36333993 | 0.31 |
ENSDART00000184379
|
CR354435.2
|
|
| chr22_+_3153876 | 0.31 |
ENSDART00000163327
|
rpl36
|
ribosomal protein L36 |
| chr22_-_15578402 | 0.30 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr25_-_35136267 | 0.30 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr4_-_18939338 | 0.30 |
ENSDART00000132081
ENSDART00000042250 |
rap1b
|
RAP1B, member of RAS oncogene family |
| chr3_-_4552590 | 0.30 |
ENSDART00000043148
ENSDART00000132224 |
ftr43
|
finTRIM family, member 43 |
| chr3_+_43086548 | 0.29 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr7_-_6429031 | 0.29 |
ENSDART00000173413
|
zgc:112234
|
zgc:112234 |
| chr15_-_1822548 | 0.29 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr3_-_45778123 | 0.29 |
ENSDART00000146211
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr3_+_27027781 | 0.29 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr3_+_57825938 | 0.29 |
ENSDART00000128815
|
cenpx
|
centromere protein X |
| chr3_-_45777226 | 0.28 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr14_-_9420911 | 0.28 |
ENSDART00000081158
|
sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr3_+_13603272 | 0.28 |
ENSDART00000185084
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr23_+_44236855 | 0.28 |
ENSDART00000130147
ENSDART00000051907 |
MEPCE
|
si:ch1073-157b13.1 |
| chr25_+_32496723 | 0.28 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr8_-_49766205 | 0.28 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr10_+_10728870 | 0.28 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr9_-_56232296 | 0.28 |
ENSDART00000149554
|
rpl31
|
ribosomal protein L31 |
| chr21_+_1647990 | 0.27 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr17_-_15189397 | 0.26 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr16_+_4770266 | 0.26 |
ENSDART00000038036
|
ube3d
|
ubiquitin protein ligase E3D |
| chr9_-_54361357 | 0.26 |
ENSDART00000149813
|
rad50
|
RAD50 homolog, double strand break repair protein |
| chr3_-_8510201 | 0.26 |
ENSDART00000009151
|
CABZ01064671.1
|
|
| chr6_-_51573975 | 0.26 |
ENSDART00000073865
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr6_+_33931740 | 0.25 |
ENSDART00000130492
ENSDART00000151213 |
orc1
|
origin recognition complex, subunit 1 |
| chr3_+_34846201 | 0.25 |
ENSDART00000055263
|
itga3a
|
integrin, alpha 3a |
| chr5_+_24047292 | 0.25 |
ENSDART00000029889
|
ctdnep1a
|
CTD nuclear envelope phosphatase 1a |
| chr25_+_3549401 | 0.25 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr8_-_20230559 | 0.24 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr23_+_29358188 | 0.24 |
ENSDART00000189242
|
tardbpl
|
TAR DNA binding protein, like |
| chr3_-_16719244 | 0.24 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr7_+_73801377 | 0.24 |
ENSDART00000184051
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr25_-_36370292 | 0.23 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr20_+_46427984 | 0.23 |
ENSDART00000060706
ENSDART00000143858 |
rad51
|
RAD51 recombinase |
| chr24_-_13349464 | 0.23 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr22_-_20419660 | 0.23 |
ENSDART00000105520
|
pias4a
|
protein inhibitor of activated STAT, 4a |
| chr7_-_6351021 | 0.23 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr3_-_14571514 | 0.22 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr5_+_41477526 | 0.22 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr22_+_24157807 | 0.22 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr6_-_42336987 | 0.22 |
ENSDART00000128777
ENSDART00000075601 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr7_-_6445129 | 0.22 |
ENSDART00000172825
|
FP325123.2
|
Histone H3.2 |
| chr6_+_22326624 | 0.22 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr2_+_32016256 | 0.21 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr7_+_55518519 | 0.21 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr2_-_57900430 | 0.21 |
ENSDART00000132245
ENSDART00000140060 |
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr20_+_13141408 | 0.21 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr25_+_8447565 | 0.21 |
ENSDART00000142090
|
fanci
|
Fanconi anemia, complementation group I |
| chr20_-_36416922 | 0.21 |
ENSDART00000019145
|
lbr
|
lamin B receptor |
| chr19_-_11846958 | 0.21 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr24_+_11908480 | 0.21 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr19_+_1873059 | 0.21 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr8_+_48848200 | 0.21 |
ENSDART00000130673
|
tp73
|
tumor protein p73 |
| chr24_+_11908833 | 0.20 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr2_+_32016516 | 0.20 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr9_-_44983666 | 0.20 |
ENSDART00000149704
|
itgb2
|
integrin, beta 2 |
| chr1_+_14020445 | 0.20 |
ENSDART00000079716
|
hpf1
|
histone PARylation factor 1 |
| chr13_+_48359573 | 0.20 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr22_-_4439311 | 0.20 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr12_-_656540 | 0.20 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr15_+_37589698 | 0.20 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr4_-_42242844 | 0.20 |
ENSDART00000163476
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr12_+_17603528 | 0.19 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr7_+_41812636 | 0.19 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr1_+_49415281 | 0.19 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr23_+_36340520 | 0.19 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr8_+_37749263 | 0.19 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr7_+_57725708 | 0.19 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr4_+_74943111 | 0.18 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr14_+_26247319 | 0.18 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr8_-_48847772 | 0.18 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr3_-_7897305 | 0.18 |
ENSDART00000169757
|
ubn2b
|
ubinuclein 2b |
| chr22_+_1911269 | 0.18 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr6_-_13200585 | 0.18 |
ENSDART00000185321
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr24_+_9693951 | 0.18 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr6_-_9282080 | 0.18 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f1_e2f7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 5.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.3 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.2 | 2.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 1.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 0.6 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 1.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.5 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 0.6 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.1 | GO:0009219 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) pyrimidine deoxyribonucleotide metabolic process(GO:0009219) |
| 0.1 | 0.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.3 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.4 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.4 | GO:0071320 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 2.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 2.0 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.7 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.4 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:1903003 | regulation of protein deubiquitination(GO:0090085) positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0090579 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 2.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 1.6 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.9 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 1.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.5 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.8 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.0 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.5 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.6 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 1.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 1.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 5.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 0.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.7 | GO:1990077 | primosome complex(GO:1990077) |
| 0.2 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 1.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 1.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.3 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.3 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.5 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 4.9 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.4 | 2.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 3.0 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.2 | 0.7 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 3.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.9 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.1 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.1 | 0.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.6 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 2.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 1.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.0 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 2.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 1.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 4.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.6 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |