Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for dharma
Z-value: 1.28
Transcription factors associated with dharma
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
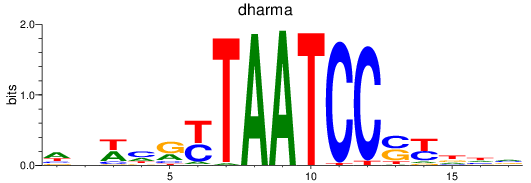
dharma
|
ENSDARG00000040955 | dharma |
Activity profile of dharma motif
Sorted Z-values of dharma motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_45177373 | 1.33 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr4_-_8030583 | 1.16 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr1_-_45146834 | 1.04 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr16_+_23403602 | 0.99 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr16_+_23404170 | 0.82 |
ENSDART00000170061
|
s100w
|
S100 calcium binding protein W |
| chr5_+_9037650 | 0.72 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr23_+_44644911 | 0.61 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr22_+_7462997 | 0.61 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr25_-_37305738 | 0.60 |
ENSDART00000154185
|
si:dkey-234i14.15
|
si:dkey-234i14.15 |
| chr10_-_45210184 | 0.58 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr19_+_7567763 | 0.55 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr4_-_77218637 | 0.52 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr14_-_25985698 | 0.51 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr8_-_25814263 | 0.50 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr7_+_8456999 | 0.46 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr22_-_6404807 | 0.44 |
ENSDART00000149399
ENSDART00000063420 |
si:rp71-1i20.2
|
si:rp71-1i20.2 |
| chr10_-_9961488 | 0.42 |
ENSDART00000191023
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr18_+_50525109 | 0.41 |
ENSDART00000098390
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr1_+_40291054 | 0.37 |
ENSDART00000136916
|
vwa10.1
|
von Willebrand factor A domain containing 10, tandem duplicate 1 |
| chr19_-_1405363 | 0.37 |
ENSDART00000165516
|
cfap20
|
cilia and flagella associated protein 20 |
| chr19_+_2631565 | 0.37 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr14_-_1667154 | 0.35 |
ENSDART00000190474
|
LO018106.1
|
|
| chr3_-_14498295 | 0.34 |
ENSDART00000172102
|
elof1
|
ELF1 homolog, elongation factor 1 |
| chr10_+_32104305 | 0.33 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr22_-_26100282 | 0.33 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr11_+_27968834 | 0.32 |
ENSDART00000147984
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr17_+_37645633 | 0.32 |
ENSDART00000085500
|
tmem229b
|
transmembrane protein 229B |
| chr16_+_19536614 | 0.31 |
ENSDART00000112894
ENSDART00000079201 ENSDART00000139357 |
sp8b
|
sp8 transcription factor b |
| chr3_+_43825785 | 0.31 |
ENSDART00000157619
|
litaf
|
lipopolysaccharide-induced TNF factor |
| chr11_+_1602916 | 0.30 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr5_-_33035295 | 0.29 |
ENSDART00000061153
|
glipr2
|
GLI pathogenesis-related 2 |
| chr2_-_32387441 | 0.29 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr10_+_11398111 | 0.29 |
ENSDART00000064210
|
cwc27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr16_+_42830152 | 0.28 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr1_+_51479128 | 0.28 |
ENSDART00000028018
|
meis1a
|
Meis homeobox 1 a |
| chr3_+_14512670 | 0.27 |
ENSDART00000161403
|
rab3db
|
RAB3D, member RAS oncogene family, b |
| chr2_+_9560740 | 0.27 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr21_-_21610857 | 0.27 |
ENSDART00000172333
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr3_+_43826018 | 0.27 |
ENSDART00000166197
ENSDART00000165145 |
litaf
|
lipopolysaccharide-induced TNF factor |
| chr7_+_62004048 | 0.27 |
ENSDART00000181818
|
smim20
|
small integral membrane protein 20 |
| chr7_-_41881177 | 0.26 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr24_+_12133814 | 0.26 |
ENSDART00000158562
ENSDART00000159029 ENSDART00000168248 |
lztfl1
|
leucine zipper transcription factor-like 1 |
| chr1_+_292545 | 0.26 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr1_-_44928987 | 0.25 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr10_-_11397590 | 0.25 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr18_+_907266 | 0.25 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr4_-_33763221 | 0.24 |
ENSDART00000132613
ENSDART00000191816 ENSDART00000188473 |
si:ch211-197f20.1
|
si:ch211-197f20.1 |
| chr16_+_42829735 | 0.23 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr15_+_15516612 | 0.23 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr5_-_69716501 | 0.23 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr15_+_21252532 | 0.22 |
ENSDART00000162619
ENSDART00000019636 ENSDART00000144901 ENSDART00000138676 ENSDART00000133821 ENSDART00000146967 ENSDART00000143990 ENSDART00000142070 ENSDART00000132373 |
usf1
|
upstream transcription factor 1 |
| chr4_-_71267777 | 0.22 |
ENSDART00000169664
|
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr22_-_26005894 | 0.22 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr4_-_76303301 | 0.22 |
ENSDART00000158504
|
zgc:174944
|
zgc:174944 |
| chr15_+_17848590 | 0.21 |
ENSDART00000168940
|
zgc:113279
|
zgc:113279 |
| chr24_+_37533728 | 0.21 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr10_+_6013076 | 0.21 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr1_-_44025066 | 0.21 |
ENSDART00000148932
|
si:ch73-109d9.1
|
si:ch73-109d9.1 |
| chr4_+_69712223 | 0.20 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr4_+_58667348 | 0.20 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr14_-_970853 | 0.20 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr3_+_26322596 | 0.19 |
ENSDART00000172700
|
si:ch211-156b7.5
|
si:ch211-156b7.5 |
| chr8_+_36142734 | 0.19 |
ENSDART00000159361
ENSDART00000161194 |
mhc2b
|
major histocompatibility complex class II integral membrane beta chain gene |
| chr4_+_76973771 | 0.19 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr23_-_46034609 | 0.19 |
ENSDART00000158712
|
zgc:65873
|
zgc:65873 |
| chr5_-_30620625 | 0.18 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr4_+_57192681 | 0.18 |
ENSDART00000190438
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr3_+_52475058 | 0.18 |
ENSDART00000035867
|
si:ch211-241f5.3
|
si:ch211-241f5.3 |
| chr4_-_45043910 | 0.18 |
ENSDART00000164068
ENSDART00000123524 ENSDART00000135010 ENSDART00000191778 |
si:ch211-227e10.6
|
si:ch211-227e10.6 |
| chr4_+_75575252 | 0.18 |
ENSDART00000166536
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr18_-_19005919 | 0.18 |
ENSDART00000129776
|
ints14
|
integrator complex subunit 14 |
| chr9_+_3170101 | 0.18 |
ENSDART00000164878
ENSDART00000160567 ENSDART00000161216 |
dync1i2a
|
dynein, cytoplasmic 1, intermediate chain 2a |
| chr4_-_71554068 | 0.18 |
ENSDART00000167414
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr4_+_71014655 | 0.17 |
ENSDART00000170837
|
si:dkeyp-80d11.14
|
si:dkeyp-80d11.14 |
| chr3_-_29891218 | 0.17 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr4_-_43616331 | 0.17 |
ENSDART00000183211
ENSDART00000150240 |
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr21_-_18086114 | 0.16 |
ENSDART00000143987
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr3_-_29891456 | 0.16 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr19_+_5640504 | 0.16 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr5_+_9147497 | 0.16 |
ENSDART00000003273
|
rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr4_-_70263609 | 0.15 |
ENSDART00000163868
|
si:dkey-3h2.3
|
si:dkey-3h2.3 |
| chr3_-_54524194 | 0.15 |
ENSDART00000155406
ENSDART00000111791 |
si:ch73-208g10.1
|
si:ch73-208g10.1 |
| chr4_-_39186893 | 0.15 |
ENSDART00000150371
ENSDART00000121804 ENSDART00000133901 |
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr21_-_19330333 | 0.15 |
ENSDART00000101462
|
mrps18c
|
mitochondrial ribosomal protein S18C |
| chr9_+_32073606 | 0.15 |
ENSDART00000184170
ENSDART00000180355 ENSDART00000110204 ENSDART00000123278 |
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr22_+_6740039 | 0.15 |
ENSDART00000144122
|
CT583625.2
|
|
| chr22_-_21314821 | 0.14 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_-_31795106 | 0.14 |
ENSDART00000164175
|
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr1_-_47122058 | 0.14 |
ENSDART00000159925
ENSDART00000101143 ENSDART00000176803 |
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr25_+_13321307 | 0.14 |
ENSDART00000161284
|
si:ch211-194m7.5
|
si:ch211-194m7.5 |
| chr4_+_966061 | 0.13 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr4_-_76105527 | 0.13 |
ENSDART00000159108
|
zgc:110171
|
zgc:110171 |
| chr5_-_22602780 | 0.13 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr18_+_5850837 | 0.13 |
ENSDART00000013150
|
cog8
|
component of oligomeric golgi complex 8 |
| chr4_-_34776147 | 0.13 |
ENSDART00000167498
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr16_-_50952266 | 0.13 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr20_-_29482492 | 0.13 |
ENSDART00000178308
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr4_-_43863085 | 0.13 |
ENSDART00000147130
|
si:dkey-261p22.1
|
si:dkey-261p22.1 |
| chr2_-_9744081 | 0.13 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr4_-_76186742 | 0.13 |
ENSDART00000163970
|
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr17_-_51260476 | 0.13 |
ENSDART00000084348
|
trappc12
|
trafficking protein particle complex 12 |
| chr18_-_17531483 | 0.12 |
ENSDART00000061000
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr4_-_75795899 | 0.12 |
ENSDART00000157925
|
si:dkey-261j11.1
|
si:dkey-261j11.1 |
| chr4_-_34480599 | 0.12 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr8_+_52642869 | 0.12 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr6_-_10863582 | 0.12 |
ENSDART00000135454
ENSDART00000133893 ENSDART00000014521 ENSDART00000164342 |
lnpk
|
lunapark, ER junction formation factor |
| chr4_-_71470455 | 0.12 |
ENSDART00000172685
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr10_-_5016997 | 0.12 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr4_-_67984478 | 0.12 |
ENSDART00000160623
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr4_-_71214516 | 0.12 |
ENSDART00000169997
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr24_+_35195963 | 0.11 |
ENSDART00000158801
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr1_+_27690 | 0.11 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr4_-_38829098 | 0.11 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr4_+_58661331 | 0.11 |
ENSDART00000188763
|
CR388148.2
|
|
| chr4_+_65487457 | 0.11 |
ENSDART00000162017
ENSDART00000193241 |
si:dkeyp-5g9.1
|
si:dkeyp-5g9.1 |
| chr4_-_47096170 | 0.11 |
ENSDART00000161079
|
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr18_+_26048128 | 0.11 |
ENSDART00000164495
ENSDART00000015712 |
znf710a
|
zinc finger protein 710a |
| chr4_+_60428812 | 0.10 |
ENSDART00000162049
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_+_1209006 | 0.10 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr5_+_4016271 | 0.10 |
ENSDART00000113627
ENSDART00000105832 ENSDART00000121415 |
ggnbp2
|
gametogenetin binding protein 2 |
| chr4_-_38828508 | 0.09 |
ENSDART00000151952
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr4_+_22480169 | 0.09 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr4_+_67994238 | 0.09 |
ENSDART00000160968
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr4_+_43225728 | 0.09 |
ENSDART00000158639
ENSDART00000141339 ENSDART00000137770 ENSDART00000166019 |
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr4_-_56673431 | 0.09 |
ENSDART00000161464
ENSDART00000191926 |
si:ch211-227p7.5
|
si:ch211-227p7.5 |
| chr4_-_56103062 | 0.09 |
ENSDART00000165784
ENSDART00000190846 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr7_+_5955160 | 0.09 |
ENSDART00000111053
|
zgc:171759
|
zgc:171759 |
| chr4_+_45389591 | 0.09 |
ENSDART00000162530
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr7_+_41146560 | 0.09 |
ENSDART00000143285
ENSDART00000173852 ENSDART00000174003 ENSDART00000038487 ENSDART00000173463 ENSDART00000166448 ENSDART00000052274 |
puf60b
|
poly-U binding splicing factor b |
| chr4_-_47095583 | 0.09 |
ENSDART00000183595
ENSDART00000183279 ENSDART00000191638 |
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr4_+_47782411 | 0.09 |
ENSDART00000161728
|
si:ch211-196h24.2
|
si:ch211-196h24.2 |
| chr4_-_43862121 | 0.08 |
ENSDART00000101961
ENSDART00000183525 |
si:dkey-261p22.1
|
si:dkey-261p22.1 |
| chr18_+_17754510 | 0.08 |
ENSDART00000189372
ENSDART00000135236 ENSDART00000137239 |
si:ch211-216l23.2
|
si:ch211-216l23.2 |
| chr4_-_52783184 | 0.08 |
ENSDART00000172283
|
si:dkey-4j21.2
|
si:dkey-4j21.2 |
| chr4_-_75651505 | 0.08 |
ENSDART00000115143
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr4_+_35227135 | 0.08 |
ENSDART00000161023
ENSDART00000182403 |
si:ch211-276i6.1
|
si:ch211-276i6.1 |
| chr13_+_40815012 | 0.08 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr7_+_17953589 | 0.08 |
ENSDART00000174778
ENSDART00000113120 |
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr4_+_36616118 | 0.08 |
ENSDART00000160058
ENSDART00000170506 |
si:dkey-151g22.1
|
si:dkey-151g22.1 |
| chr16_+_29586004 | 0.08 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr4_-_33073382 | 0.08 |
ENSDART00000146414
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr3_+_23092762 | 0.08 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr4_-_38539463 | 0.07 |
ENSDART00000158211
|
si:ch211-209n20.3
|
si:ch211-209n20.3 |
| chr18_+_21951551 | 0.07 |
ENSDART00000146261
|
ranbp10
|
RAN binding protein 10 |
| chr23_-_41824460 | 0.07 |
ENSDART00000131235
|
GMEB2
|
si:ch73-302a13.2 |
| chr4_-_34459615 | 0.07 |
ENSDART00000169144
ENSDART00000167022 |
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr16_-_21915223 | 0.07 |
ENSDART00000170634
|
setdb1b
|
SET domain, bifurcated 1b |
| chr3_-_56030117 | 0.07 |
ENSDART00000113030
ENSDART00000157764 |
cep112
|
centrosomal protein 112 |
| chr21_+_5080789 | 0.07 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr17_-_30944566 | 0.07 |
ENSDART00000192663
|
evla
|
Enah/Vasp-like a |
| chr11_-_7355995 | 0.07 |
ENSDART00000156595
|
si:dkey-197i20.6
|
si:dkey-197i20.6 |
| chr4_+_46812891 | 0.07 |
ENSDART00000189655
ENSDART00000162329 |
si:ch211-215p11.3
|
si:ch211-215p11.3 |
| chr19_+_3140313 | 0.07 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr10_+_2841205 | 0.06 |
ENSDART00000131505
ENSDART00000055869 |
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr4_-_63559311 | 0.06 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr14_+_45676701 | 0.06 |
ENSDART00000183505
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr2_-_5356686 | 0.06 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr5_-_22602979 | 0.06 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr24_-_24060632 | 0.06 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr6_+_41181869 | 0.06 |
ENSDART00000002046
|
opn1mw1
|
opsin 1 (cone pigments), medium-wave-sensitive, 1 |
| chr4_+_45388995 | 0.06 |
ENSDART00000150802
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr18_+_7639401 | 0.05 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr21_-_20342096 | 0.05 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr15_-_8191841 | 0.05 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr9_+_3548529 | 0.05 |
ENSDART00000130861
ENSDART00000123074 ENSDART00000147650 |
tlk1a
|
tousled-like kinase 1a |
| chr16_+_14201401 | 0.05 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr11_-_21585176 | 0.04 |
ENSDART00000045391
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr17_-_33414781 | 0.04 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr3_+_57268363 | 0.04 |
ENSDART00000180725
|
TMEM235 (1 of many)
|
transmembrane protein 235 |
| chr15_-_568645 | 0.04 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr3_-_55128258 | 0.03 |
ENSDART00000101734
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr4_+_40383229 | 0.03 |
ENSDART00000126906
ENSDART00000191198 |
si:ch211-218h8.1
|
si:ch211-218h8.1 |
| chr9_-_6502491 | 0.03 |
ENSDART00000102672
|
nck2a
|
NCK adaptor protein 2a |
| chr7_+_66884570 | 0.03 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr4_-_63558838 | 0.02 |
ENSDART00000191708
ENSDART00000190740 ENSDART00000186476 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr21_+_44773237 | 0.02 |
ENSDART00000084780
|
rnf121
|
ring finger protein 121 |
| chr21_-_20341836 | 0.02 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr11_+_20056732 | 0.02 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr4_+_54862025 | 0.02 |
ENSDART00000166721
|
si:dkey-56m15.9
|
si:dkey-56m15.9 |
| chr5_+_13394543 | 0.02 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr4_+_70341246 | 0.02 |
ENSDART00000123408
|
si:ch211-76m11.3
|
si:ch211-76m11.3 |
| chr25_-_35121946 | 0.02 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr4_+_51201875 | 0.02 |
ENSDART00000189875
ENSDART00000186722 |
BX005380.1
|
|
| chr11_+_25157374 | 0.02 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr8_+_30747940 | 0.02 |
ENSDART00000098975
|
p2rx4b
|
purinergic receptor P2X, ligand-gated ion channel, 4b |
| chr4_-_34460301 | 0.01 |
ENSDART00000192977
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr14_-_7141550 | 0.01 |
ENSDART00000172020
|
trpt1
|
tRNA phosphotransferase 1 |
| chr8_+_46327350 | 0.01 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr20_+_2139436 | 0.00 |
ENSDART00000155311
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr15_-_8191992 | 0.00 |
ENSDART00000155381
ENSDART00000190714 |
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr5_-_33022014 | 0.00 |
ENSDART00000061149
|
zgc:55461
|
zgc:55461 |
| chr18_+_19006063 | 0.00 |
ENSDART00000135729
|
slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr12_-_19185865 | 0.00 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr23_+_20705849 | 0.00 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dharma
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.5 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.2 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.5 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.3 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.3 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.0 | 0.1 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.6 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.3 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.8 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 2.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |