Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
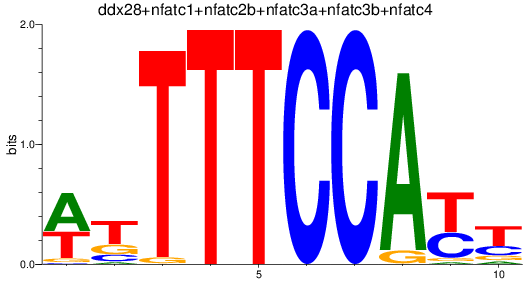
Results for ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Z-value: 1.76
Transcription factors associated with ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc1
|
ENSDARG00000036168 | nuclear factor of activated T cells 1 |
|
nfatc3b
|
ENSDARG00000051729 | nuclear factor of activated T cells 3b |
|
nfatc4
|
ENSDARG00000054162 | nuclear factor of activated T cells 4 |
|
nfatc3a
|
ENSDARG00000076297 | nuclear factor of activated T cells 3a |
|
nfatc2b
|
ENSDARG00000079972 | nuclear factor of activated T cells 2b |
|
ddx28
|
ENSDARG00000112133 | nuclear factor of activated T cells 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc1 | dr11_v1_chr19_+_22216778_22216778 | 0.94 | 1.9e-02 | Click! |
| nfatc2b | dr11_v1_chr6_-_55254786_55254786 | -0.39 | 5.1e-01 | Click! |
| nfatc3b | dr11_v1_chr25_-_36248053_36248053 | 0.32 | 5.9e-01 | Click! |
| nfatc3a | dr11_v1_chr7_-_34927961_34927961 | 0.30 | 6.3e-01 | Click! |
| nfatc4 | dr11_v1_chr2_-_37797577_37797577 | 0.13 | 8.3e-01 | Click! |
Activity profile of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
Sorted Z-values of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_26064480 | 1.46 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr13_+_23157053 | 1.45 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr15_-_20933574 | 1.17 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr18_+_26743749 | 1.13 |
ENSDART00000145212
|
alpk3a
|
alpha-kinase 3a |
| chr1_-_38816685 | 1.09 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr2_-_24289641 | 0.97 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr6_-_10233538 | 0.95 |
ENSDART00000182004
ENSDART00000149237 ENSDART00000148876 |
xirp2a
|
xin actin binding repeat containing 2a |
| chr22_-_15587360 | 0.89 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr14_+_49296052 | 0.86 |
ENSDART00000006073
ENSDART00000105346 |
anxa6
|
annexin A6 |
| chr3_-_39152478 | 0.85 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr9_+_6587056 | 0.83 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr9_+_6587364 | 0.82 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr8_-_40464935 | 0.80 |
ENSDART00000040013
|
myl7
|
myosin, light chain 7, regulatory |
| chr11_-_29946640 | 0.80 |
ENSDART00000079175
|
zgc:113276
|
zgc:113276 |
| chr9_-_42989297 | 0.80 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr1_-_41192059 | 0.79 |
ENSDART00000084665
ENSDART00000135369 |
dok7
|
docking protein 7 |
| chr1_-_38195012 | 0.79 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr25_-_28674739 | 0.78 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr19_+_5674907 | 0.74 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr3_+_14463941 | 0.72 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr13_+_35690023 | 0.72 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr2_+_42191592 | 0.72 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr12_+_17106117 | 0.70 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr7_+_65564163 | 0.68 |
ENSDART00000082679
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr11_-_30611814 | 0.66 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr2_-_24369087 | 0.65 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr11_-_30612166 | 0.64 |
ENSDART00000177984
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr25_+_6122823 | 0.62 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr16_+_25245857 | 0.60 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr3_-_32817274 | 0.57 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr12_+_16281312 | 0.56 |
ENSDART00000152500
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr13_-_22646134 | 0.56 |
ENSDART00000136808
|
mmrn2a
|
multimerin 2a |
| chr4_+_2620751 | 0.55 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr2_+_4207209 | 0.54 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr20_+_1996202 | 0.54 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr7_-_21905851 | 0.53 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr14_-_33454595 | 0.52 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr7_-_52558495 | 0.51 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr7_-_52531252 | 0.51 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr23_+_13814978 | 0.51 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr18_+_26750516 | 0.50 |
ENSDART00000110843
|
alpk3a
|
alpha-kinase 3a |
| chr13_-_13754091 | 0.50 |
ENSDART00000131255
|
ky
|
kyphoscoliosis peptidase |
| chr11_-_30601401 | 0.50 |
ENSDART00000172244
ENSDART00000046800 |
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr5_+_30179010 | 0.50 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| chr16_-_12723324 | 0.50 |
ENSDART00000131915
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr21_+_29077509 | 0.48 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr20_+_53577502 | 0.48 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr3_-_61181018 | 0.47 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr19_-_41069573 | 0.47 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr16_+_30438041 | 0.47 |
ENSDART00000137977
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr18_+_26719787 | 0.46 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr6_-_50204262 | 0.45 |
ENSDART00000163648
|
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr6_-_36552844 | 0.45 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr24_+_35975398 | 0.45 |
ENSDART00000173058
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr7_-_30367650 | 0.44 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr25_+_14087045 | 0.44 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr25_-_32888115 | 0.43 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr8_+_27743550 | 0.42 |
ENSDART00000046004
|
wnt2bb
|
wingless-type MMTV integration site family, member 2Bb |
| chr21_+_21621042 | 0.42 |
ENSDART00000134907
|
tgfb1b
|
transforming growth factor, beta 1b |
| chr18_+_5273953 | 0.42 |
ENSDART00000165073
|
slc12a1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr7_-_52417777 | 0.41 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr20_+_31287356 | 0.40 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr9_+_31752628 | 0.39 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr6_-_48094342 | 0.39 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr8_-_19342111 | 0.39 |
ENSDART00000138881
|
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr10_+_33990572 | 0.38 |
ENSDART00000138547
|
fryb
|
furry homolog b (Drosophila) |
| chr7_+_44445595 | 0.38 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr9_+_33145522 | 0.37 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr20_+_23173710 | 0.37 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr22_+_28446557 | 0.36 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr10_-_7892192 | 0.36 |
ENSDART00000145480
|
smtna
|
smoothelin a |
| chr9_-_34944604 | 0.36 |
ENSDART00000140563
ENSDART00000136812 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr11_-_34628789 | 0.35 |
ENSDART00000192433
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr10_-_42685512 | 0.35 |
ENSDART00000081347
|
stc1l
|
stanniocalcin 1, like |
| chr6_+_48978202 | 0.35 |
ENSDART00000150023
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr22_+_28446365 | 0.35 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr21_-_22928214 | 0.35 |
ENSDART00000182760
|
dub
|
duboraya |
| chr7_+_27059330 | 0.34 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr6_-_33875919 | 0.33 |
ENSDART00000190411
|
tmem69
|
transmembrane protein 69 |
| chr8_+_17078692 | 0.31 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr11_-_11471857 | 0.30 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr19_+_43392446 | 0.30 |
ENSDART00000147290
|
yrk
|
Yes-related kinase |
| chr13_-_39947335 | 0.30 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr3_-_20091964 | 0.29 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr25_-_12902242 | 0.29 |
ENSDART00000164733
|
sept15
|
septin 15 |
| chr8_-_45834825 | 0.29 |
ENSDART00000132965
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr2_+_38161318 | 0.28 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr15_-_1066269 | 0.28 |
ENSDART00000093133
|
aadac
|
arylacetamide deacetylase |
| chr12_-_15205087 | 0.28 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr21_-_17603182 | 0.28 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr9_+_4429593 | 0.28 |
ENSDART00000184855
|
FP015810.1
|
|
| chr9_-_23990416 | 0.27 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr13_-_18835254 | 0.27 |
ENSDART00000147579
ENSDART00000146795 |
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr14_-_3268155 | 0.27 |
ENSDART00000177244
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr16_-_35975254 | 0.27 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr20_+_17783404 | 0.27 |
ENSDART00000181946
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr2_-_25140022 | 0.26 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr4_-_9722568 | 0.26 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr23_-_19682971 | 0.26 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr13_+_1575276 | 0.26 |
ENSDART00000165987
|
DST
|
dystonin |
| chr13_+_49175624 | 0.26 |
ENSDART00000193550
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr22_-_3564563 | 0.26 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr25_-_14087377 | 0.25 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr23_+_27703749 | 0.25 |
ENSDART00000027224
|
lmbr1l
|
limb development membrane protein 1-like |
| chr10_+_8437930 | 0.25 |
ENSDART00000074553
|
pptc7b
|
PTC7 protein phosphatase homolog b |
| chr4_-_10826575 | 0.25 |
ENSDART00000164771
ENSDART00000067256 |
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr11_+_13630107 | 0.25 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr4_+_7677318 | 0.25 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr10_-_28761454 | 0.23 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr10_-_24689280 | 0.23 |
ENSDART00000191476
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr13_-_42749916 | 0.23 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr4_-_49607096 | 0.23 |
ENSDART00000154381
|
si:dkey-159n16.4
|
si:dkey-159n16.4 |
| chr16_-_13613475 | 0.23 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr17_+_12865746 | 0.23 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr12_+_27243059 | 0.23 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr11_+_10541258 | 0.23 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr15_-_25099233 | 0.23 |
ENSDART00000192617
|
rflnb
|
refilin B |
| chr15_-_34468599 | 0.22 |
ENSDART00000192984
|
meox2a
|
mesenchyme homeobox 2a |
| chr19_-_33087246 | 0.22 |
ENSDART00000052078
|
klhl38a
|
kelch-like family member 38a |
| chr5_-_42950963 | 0.22 |
ENSDART00000149868
|
grsf1
|
G-rich RNA sequence binding factor 1 |
| chr21_-_43131752 | 0.22 |
ENSDART00000024137
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr6_+_27992886 | 0.22 |
ENSDART00000160354
|
amotl2a
|
angiomotin like 2a |
| chr16_+_46430627 | 0.22 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr6_+_49095646 | 0.22 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr14_+_26759332 | 0.22 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr3_+_39099716 | 0.21 |
ENSDART00000083388
|
tmem98
|
transmembrane protein 98 |
| chr18_+_17786548 | 0.21 |
ENSDART00000189493
ENSDART00000146133 |
ZNF423
|
si:ch211-216l23.1 |
| chr19_+_26340736 | 0.21 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr5_-_48307804 | 0.21 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr15_-_25099679 | 0.21 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr16_+_23398369 | 0.21 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr11_-_40165683 | 0.21 |
ENSDART00000186510
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr23_+_27740788 | 0.21 |
ENSDART00000053871
|
dhh
|
desert hedgehog |
| chr14_-_448182 | 0.20 |
ENSDART00000180018
|
FAT4
|
FAT atypical cadherin 4 |
| chr2_-_3611960 | 0.20 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr9_-_35069645 | 0.20 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr15_-_25518084 | 0.20 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr16_+_31542645 | 0.19 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr10_+_8767541 | 0.19 |
ENSDART00000170272
|
itga1
|
integrin, alpha 1 |
| chr5_-_68074592 | 0.19 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr23_+_36101185 | 0.19 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr5_+_6617401 | 0.19 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr6_-_49673476 | 0.19 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr13_+_49175947 | 0.19 |
ENSDART00000056927
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr21_+_2091895 | 0.19 |
ENSDART00000161828
|
si:rp71-1h20.9
|
si:rp71-1h20.9 |
| chr19_-_38611814 | 0.19 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr6_+_60112200 | 0.19 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr25_-_12635371 | 0.19 |
ENSDART00000162463
|
zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr8_-_38403018 | 0.18 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr16_+_35887868 | 0.18 |
ENSDART00000169677
ENSDART00000170772 |
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr25_+_35683956 | 0.18 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr20_+_32118559 | 0.18 |
ENSDART00000026273
|
cd164
|
CD164 molecule, sialomucin |
| chr6_+_52263236 | 0.18 |
ENSDART00000144174
|
esyt1b
|
extended synaptotagmin-like protein 1b |
| chr25_-_35524166 | 0.18 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr13_+_16279890 | 0.18 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr15_-_26923993 | 0.18 |
ENSDART00000132459
|
ccdc9
|
coiled-coil domain containing 9 |
| chr23_+_23119008 | 0.17 |
ENSDART00000132418
|
samd11
|
sterile alpha motif domain containing 11 |
| chr5_-_23277939 | 0.17 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr13_-_31470439 | 0.17 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr20_-_47051996 | 0.17 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr9_-_10805231 | 0.17 |
ENSDART00000193913
ENSDART00000078348 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr4_+_76926059 | 0.17 |
ENSDART00000136192
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr13_-_12581388 | 0.17 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr9_-_12424791 | 0.17 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr7_+_52761841 | 0.17 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr8_-_19904124 | 0.16 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr5_-_24270989 | 0.16 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr9_+_27876146 | 0.16 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr16_+_23397785 | 0.16 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr12_+_46960579 | 0.16 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr19_+_19652439 | 0.16 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr1_-_40102836 | 0.16 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr9_+_43799829 | 0.16 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr11_-_24532988 | 0.16 |
ENSDART00000067078
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr18_+_17786710 | 0.16 |
ENSDART00000190203
ENSDART00000187095 ENSDART00000083296 |
ZNF423
|
si:ch211-216l23.1 |
| chr10_-_43771447 | 0.16 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr7_-_38658411 | 0.16 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr25_-_16818978 | 0.16 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr16_+_40301056 | 0.15 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr1_-_58664854 | 0.15 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr12_+_20627505 | 0.15 |
ENSDART00000074384
|
stx4
|
syntaxin 4 |
| chr8_+_2656681 | 0.15 |
ENSDART00000185067
ENSDART00000165943 |
fam102aa
|
family with sequence similarity 102, member Aa |
| chr9_-_23994225 | 0.15 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr13_+_13681681 | 0.15 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr8_-_49728590 | 0.15 |
ENSDART00000135714
ENSDART00000138810 ENSDART00000098319 |
gkap1
|
G kinase anchoring protein 1 |
| chr2_+_10914023 | 0.15 |
ENSDART00000146546
|
mrpl37
|
mitochondrial ribosomal protein L37 |
| chr20_+_6773790 | 0.15 |
ENSDART00000169966
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr20_-_31496679 | 0.14 |
ENSDART00000153437
ENSDART00000153193 |
sash1a
|
SAM and SH3 domain containing 1a |
| chr5_-_44496553 | 0.14 |
ENSDART00000178081
|
gas1a
|
growth arrest-specific 1a |
| chr15_-_21852963 | 0.14 |
ENSDART00000153581
ENSDART00000155588 ENSDART00000079399 |
laynb
|
layilin b |
| chr13_-_377256 | 0.14 |
ENSDART00000137398
|
ch1073-291c23.1
|
ch1073-291c23.1 |
| chr9_-_16133263 | 0.14 |
ENSDART00000077187
|
myo1b
|
myosin IB |
| chr17_-_42799104 | 0.13 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr17_+_30448452 | 0.13 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr1_+_36772348 | 0.13 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr13_+_43639867 | 0.13 |
ENSDART00000042588
ENSDART00000074728 |
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr18_+_39416357 | 0.13 |
ENSDART00000183174
ENSDART00000127955 ENSDART00000171303 |
lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr9_-_8454060 | 0.13 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr9_+_25832329 | 0.13 |
ENSDART00000130059
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr24_-_21819010 | 0.13 |
ENSDART00000091096
|
CR352265.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.9 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 1.3 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.4 | GO:1900158 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.8 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.6 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.7 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.5 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.3 | GO:0021577 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.1 | 1.8 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.5 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.4 | GO:0001955 | blood vessel maturation(GO:0001955) regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.8 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 0.3 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.5 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.1 | GO:1904407 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.1 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.0 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0035912 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 2.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.6 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.2 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.2 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.0 | 0.1 | GO:0060406 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0071456 | response to activity(GO:0014823) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 1.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |