Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
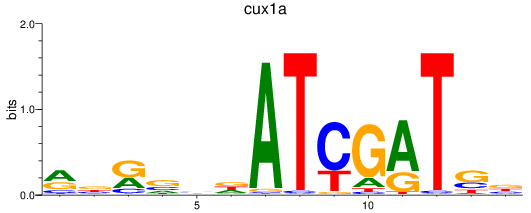
Results for cux1a
Z-value: 0.21
Transcription factors associated with cux1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cux1a
|
ENSDARG00000078459 | cut-like homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cux1a | dr11_v1_chr10_-_33156789_33156789 | 0.88 | 4.9e-02 | Click! |
Activity profile of cux1a motif
Sorted Z-values of cux1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_35334642 | 0.18 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr25_+_21324588 | 0.17 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr23_-_33986762 | 0.14 |
ENSDART00000144609
|
tmed4
|
transmembrane p24 trafficking protein 4 |
| chr4_-_25485404 | 0.13 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr18_-_40481028 | 0.13 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr6_-_29105727 | 0.13 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr17_+_45454943 | 0.12 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr18_+_45571378 | 0.11 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr20_-_18736281 | 0.11 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr12_-_7607114 | 0.11 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr25_-_6082509 | 0.11 |
ENSDART00000104755
|
cpeb1a
|
cytoplasmic polyadenylation element binding protein 1a |
| chr12_+_10115964 | 0.11 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr2_-_37353098 | 0.10 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr22_-_15562933 | 0.10 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr10_+_21786656 | 0.10 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr23_-_44494401 | 0.09 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr2_-_37352514 | 0.09 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr10_+_34377697 | 0.09 |
ENSDART00000189441
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr17_+_11507131 | 0.09 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr1_-_39976492 | 0.09 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr14_-_33083539 | 0.09 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr3_-_38692920 | 0.08 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr7_+_29951997 | 0.08 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr11_+_41089574 | 0.07 |
ENSDART00000170023
|
phf13
|
PHD finger protein 13 |
| chr14_-_32486757 | 0.07 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr15_-_9419472 | 0.07 |
ENSDART00000122117
|
sacs
|
sacsin molecular chaperone |
| chr19_+_14059349 | 0.07 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr15_-_16704417 | 0.07 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr13_-_36264861 | 0.06 |
ENSDART00000100204
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr12_+_38770654 | 0.06 |
ENSDART00000155367
|
kif19
|
kinesin family member 19 |
| chr7_+_49272702 | 0.06 |
ENSDART00000083389
|
abtb2a
|
ankyrin repeat and BTB (POZ) domain containing 2a |
| chr7_-_25895189 | 0.06 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr15_+_36966369 | 0.06 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr14_-_25985698 | 0.06 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr10_+_44057177 | 0.06 |
ENSDART00000164610
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr7_-_12789251 | 0.05 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr8_+_16758304 | 0.05 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr7_+_29863548 | 0.05 |
ENSDART00000136555
ENSDART00000134068 |
tln2a
|
talin 2a |
| chr8_-_24135171 | 0.05 |
ENSDART00000112926
|
adora1b
|
adenosine A1 receptor b |
| chr2_-_32513538 | 0.04 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr8_-_19451487 | 0.04 |
ENSDART00000184037
|
zgc:92140
|
zgc:92140 |
| chr16_+_39242339 | 0.04 |
ENSDART00000102510
|
zgc:77056
|
zgc:77056 |
| chr16_-_25380903 | 0.04 |
ENSDART00000086375
ENSDART00000188587 |
adnp2a
|
ADNP homeobox 2a |
| chr12_+_14149686 | 0.04 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr15_+_42626957 | 0.04 |
ENSDART00000154754
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr20_-_54075136 | 0.04 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr6_+_38896158 | 0.04 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr2_+_7715810 | 0.04 |
ENSDART00000130781
|
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr24_+_35881124 | 0.04 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr21_-_26691959 | 0.03 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_+_16152316 | 0.03 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr16_-_24561354 | 0.03 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr5_-_13835461 | 0.03 |
ENSDART00000148297
ENSDART00000114841 |
add2
|
adducin 2 (beta) |
| chr23_+_3721042 | 0.03 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr8_-_29851706 | 0.02 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr24_-_3407507 | 0.02 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr23_+_9353552 | 0.02 |
ENSDART00000163298
|
BX511246.1
|
|
| chr7_-_56766973 | 0.02 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr21_-_20120543 | 0.02 |
ENSDART00000065664
|
dusp4
|
dual specificity phosphatase 4 |
| chr17_+_24597001 | 0.01 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr12_-_9700605 | 0.01 |
ENSDART00000161063
|
heatr1
|
HEAT repeat containing 1 |
| chr22_-_25078650 | 0.01 |
ENSDART00000131811
ENSDART00000141546 |
kat14
|
lysine acetyltransferase 14 |
| chr12_-_19091214 | 0.01 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr11_-_42396302 | 0.01 |
ENSDART00000165624
|
slmapa
|
sarcolemma associated protein a |
| chr2_-_56649883 | 0.01 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr22_+_21516689 | 0.00 |
ENSDART00000105550
ENSDART00000136374 |
mier2
|
mesoderm induction early response 1, family member 2 |
| chr23_-_24047054 | 0.00 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr11_-_16152105 | 0.00 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr11_-_16152400 | 0.00 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of cux1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |