Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
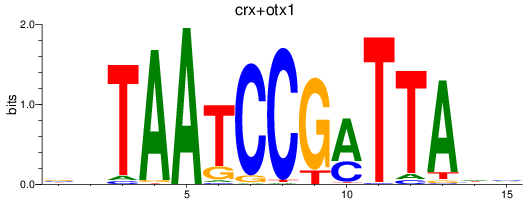
Results for crx+otx1
Z-value: 0.95
Transcription factors associated with crx+otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
crx
|
ENSDARG00000011989 | cone-rod homeobox |
|
otx1
|
ENSDARG00000094992 | orthodenticle homeobox 1 |
|
crx
|
ENSDARG00000113850 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx1 | dr11_v1_chr17_+_24318753_24318753 | 0.32 | 6.0e-01 | Click! |
| crx | dr11_v1_chr5_+_36932718_36932718 | -0.22 | 7.2e-01 | Click! |
Activity profile of crx+otx1 motif
Sorted Z-values of crx+otx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_3366782 | 0.53 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr10_+_17026870 | 0.47 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr16_-_46579936 | 0.46 |
ENSDART00000166143
ENSDART00000127212 |
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr15_+_46605482 | 0.41 |
ENSDART00000184449
|
CABZ01044048.1
|
|
| chr24_-_21689146 | 0.39 |
ENSDART00000105917
|
urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr16_-_16701718 | 0.37 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr13_+_733027 | 0.37 |
ENSDART00000149547
|
nrxn1b
|
neurexin 1b |
| chr9_-_3519253 | 0.34 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr22_-_16154771 | 0.34 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr22_+_25453334 | 0.33 |
ENSDART00000123962
|
si:ch211-12h2.6
|
si:ch211-12h2.6 |
| chr9_-_38042823 | 0.32 |
ENSDART00000141998
ENSDART00000133998 |
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr4_+_72797711 | 0.31 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr2_-_51757328 | 0.31 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr6_-_58757131 | 0.29 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr20_-_54014373 | 0.28 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr8_-_46572298 | 0.27 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr20_-_54014539 | 0.25 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr5_-_69180587 | 0.24 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr17_+_30448452 | 0.22 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr13_+_41917606 | 0.22 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr18_-_10713414 | 0.22 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr11_-_1550709 | 0.21 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr7_-_26518086 | 0.20 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr5_-_69180227 | 0.19 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr21_+_43702016 | 0.19 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr7_+_20030888 | 0.19 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr21_+_31838386 | 0.19 |
ENSDART00000135591
|
si:ch211-12m10.1
|
si:ch211-12m10.1 |
| chr17_-_45125537 | 0.18 |
ENSDART00000113552
|
zgc:163014
|
zgc:163014 |
| chr11_+_19080400 | 0.18 |
ENSDART00000044423
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr7_+_13702248 | 0.18 |
ENSDART00000172811
|
pdcd7
|
programmed cell death 7 |
| chr10_-_42898220 | 0.18 |
ENSDART00000099270
|
CU326366.2
|
|
| chr10_+_4875262 | 0.18 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr9_-_394088 | 0.17 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr24_-_38079261 | 0.16 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr2_+_44972720 | 0.16 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr1_-_43712120 | 0.16 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr3_-_12903609 | 0.16 |
ENSDART00000160522
|
ccdc137
|
coiled-coil domain containing 137 |
| chr11_+_18873113 | 0.15 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr4_+_68562464 | 0.15 |
ENSDART00000192954
|
BX548011.4
|
|
| chr25_+_7532627 | 0.15 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr12_-_35883814 | 0.15 |
ENSDART00000177986
ENSDART00000129888 |
cep131
|
centrosomal protein 131 |
| chr14_-_6402769 | 0.15 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr23_+_7710721 | 0.15 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr9_+_3519191 | 0.14 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr1_-_24349759 | 0.14 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr16_+_38159758 | 0.14 |
ENSDART00000058666
ENSDART00000112165 |
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr3_-_15679107 | 0.14 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr13_-_30161684 | 0.14 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| chr19_-_5207361 | 0.13 |
ENSDART00000174611
|
stx1b
|
syntaxin 1B |
| chr18_-_8030073 | 0.13 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr11_-_37693019 | 0.13 |
ENSDART00000102898
|
zgc:158258
|
zgc:158258 |
| chr21_-_28340977 | 0.13 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr20_+_13969414 | 0.12 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr19_+_2876106 | 0.12 |
ENSDART00000189309
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr2_-_42864472 | 0.12 |
ENSDART00000134139
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr24_+_20536056 | 0.12 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr7_+_22330466 | 0.11 |
ENSDART00000187347
ENSDART00000174483 |
fgf11a
|
fibroblast growth factor 11a |
| chr16_+_43344475 | 0.11 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr25_+_35913614 | 0.11 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr2_-_31686353 | 0.10 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr14_+_11909966 | 0.10 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr20_+_46560258 | 0.10 |
ENSDART00000122115
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr18_+_3634652 | 0.10 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr9_+_21151138 | 0.10 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr20_-_10120442 | 0.10 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr13_-_29406534 | 0.10 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr10_-_25852517 | 0.10 |
ENSDART00000191551
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr19_+_45970692 | 0.09 |
ENSDART00000158781
|
si:ch211-153f2.7
|
si:ch211-153f2.7 |
| chr14_+_52413846 | 0.09 |
ENSDART00000160952
|
noa1
|
nitric oxide associated 1 |
| chr2_-_15349382 | 0.09 |
ENSDART00000057238
|
olfm3b
|
olfactomedin 3b |
| chr7_+_67486807 | 0.09 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr8_+_28724692 | 0.09 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr10_-_40968095 | 0.09 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr16_+_43219363 | 0.09 |
ENSDART00000183610
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr23_+_30827959 | 0.09 |
ENSDART00000033429
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr10_-_322769 | 0.09 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr13_-_40499296 | 0.09 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr8_+_2656231 | 0.09 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr5_+_66355153 | 0.09 |
ENSDART00000082745
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr4_+_33461796 | 0.08 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr25_-_19090479 | 0.08 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr11_+_6295370 | 0.08 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr25_+_21324588 | 0.08 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr19_-_3106447 | 0.08 |
ENSDART00000137987
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr2_+_24080694 | 0.08 |
ENSDART00000024058
|
kcnh2a
|
potassium voltage-gated channel, subfamily H (eag-related), member 2a |
| chr5_+_9434288 | 0.08 |
ENSDART00000162089
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr6_-_10168822 | 0.08 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr21_-_20765338 | 0.08 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr8_+_18545933 | 0.08 |
ENSDART00000148806
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr5_-_23715027 | 0.08 |
ENSDART00000139020
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr21_-_44104600 | 0.08 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr1_-_58766424 | 0.08 |
ENSDART00000191010
|
CABZ01096573.1
|
|
| chr2_-_39036604 | 0.08 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr12_-_33659328 | 0.08 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr19_+_14921000 | 0.08 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr5_-_27993972 | 0.07 |
ENSDART00000175819
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr4_-_28353538 | 0.07 |
ENSDART00000064219
|
trmu
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr5_-_65082107 | 0.07 |
ENSDART00000162511
|
inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr17_+_50701748 | 0.07 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr13_+_2442841 | 0.07 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr2_-_26499842 | 0.07 |
ENSDART00000186929
|
BX569796.1
|
|
| chr20_+_1272526 | 0.07 |
ENSDART00000008115
ENSDART00000133825 |
hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr14_-_31694274 | 0.07 |
ENSDART00000173353
|
map7d3
|
MAP7 domain containing 3 |
| chr7_-_39378903 | 0.07 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr4_-_17353100 | 0.07 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr10_+_39476600 | 0.07 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr9_+_17982737 | 0.07 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr13_+_31002658 | 0.07 |
ENSDART00000140257
|
lrrc18a
|
leucine rich repeat containing 18a |
| chr6_+_54538948 | 0.07 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr21_-_2209012 | 0.07 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr19_+_7152966 | 0.07 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr23_-_42876596 | 0.07 |
ENSDART00000086156
|
dlgap4a
|
discs, large (Drosophila) homolog-associated protein 4a |
| chr5_+_26847190 | 0.07 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr9_-_46276626 | 0.07 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr3_+_15296824 | 0.06 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr14_-_4273396 | 0.06 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr25_-_4733008 | 0.06 |
ENSDART00000191884
|
drd4a
|
dopamine receptor D4a |
| chr10_+_39476432 | 0.06 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr24_+_10039165 | 0.06 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr8_+_31435452 | 0.06 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr18_-_6975175 | 0.06 |
ENSDART00000134194
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr2_+_49417900 | 0.06 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr16_-_16225260 | 0.06 |
ENSDART00000165790
|
gra
|
granulito |
| chr3_-_26183699 | 0.06 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr9_+_19095023 | 0.06 |
ENSDART00000110457
ENSDART00000099426 ENSDART00000137087 |
crfb1
|
cytokine receptor family member b1 |
| chr22_+_1170294 | 0.06 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr7_+_39011852 | 0.06 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr7_+_19903924 | 0.06 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr4_-_36522557 | 0.06 |
ENSDART00000167619
|
BX649453.1
|
Danio rerio gastrula zinc finger protein XlCGF8.2DB-like (LOC100150619), mRNA. |
| chr25_+_35891342 | 0.06 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr1_+_23162124 | 0.06 |
ENSDART00000188428
|
si:dkey-92j12.5
|
si:dkey-92j12.5 |
| chr17_+_31237866 | 0.06 |
ENSDART00000076943
|
prlh2r
|
prolactin releasing hormone 2 receptor |
| chr2_+_33051730 | 0.06 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr5_+_11407504 | 0.06 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr19_+_34169055 | 0.06 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr5_-_23715861 | 0.06 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr17_+_10593398 | 0.06 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr11_-_41996957 | 0.06 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr22_+_17203752 | 0.06 |
ENSDART00000143376
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr5_+_51833305 | 0.05 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr2_-_37537887 | 0.05 |
ENSDART00000143496
ENSDART00000025841 |
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr7_+_19904136 | 0.05 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr24_-_31140356 | 0.05 |
ENSDART00000167837
|
tmem56a
|
transmembrane protein 56a |
| chr21_-_39058490 | 0.05 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr4_+_36083288 | 0.05 |
ENSDART00000172017
|
si:ch211-271g18.4
|
si:ch211-271g18.4 |
| chr3_+_22999849 | 0.05 |
ENSDART00000127239
|
si:ch211-237i5.4
|
si:ch211-237i5.4 |
| chr9_-_8314028 | 0.05 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr16_+_9726038 | 0.05 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr24_-_25098719 | 0.05 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr21_+_33503835 | 0.05 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr4_+_12111154 | 0.05 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr20_+_18260358 | 0.05 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_-_25028527 | 0.05 |
ENSDART00000040733
|
cd209
|
CD209 molecule |
| chr23_+_1181248 | 0.05 |
ENSDART00000170942
|
utrn
|
utrophin |
| chr2_+_33052169 | 0.05 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr18_+_27439680 | 0.05 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr13_+_6189203 | 0.05 |
ENSDART00000109665
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr25_+_32031536 | 0.05 |
ENSDART00000173656
|
tjp1b
|
tight junction protein 1b |
| chr3_+_25508247 | 0.05 |
ENSDART00000039448
|
mchr1b
|
melanin-concentrating hormone receptor 1b |
| chr15_-_18213515 | 0.05 |
ENSDART00000101635
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr3_+_50602866 | 0.05 |
ENSDART00000190069
ENSDART00000153921 |
gsg1l2a
|
gsg1-like 2a |
| chr2_+_20430366 | 0.05 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr10_+_40235959 | 0.05 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr25_-_10630496 | 0.05 |
ENSDART00000153639
ENSDART00000181722 ENSDART00000177834 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr17_-_7371564 | 0.05 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr4_+_44808133 | 0.04 |
ENSDART00000184430
|
znf1107
|
zinc finger protein 1107 |
| chr21_+_36581384 | 0.04 |
ENSDART00000183924
|
BX936439.1
|
|
| chr4_-_908821 | 0.04 |
ENSDART00000168266
|
mrap2b
|
melanocortin 2 receptor accessory protein 2b |
| chr3_+_32492467 | 0.04 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr21_-_40317035 | 0.04 |
ENSDART00000143648
ENSDART00000013359 |
or101-1
|
odorant receptor, family B, subfamily 101, member 1 |
| chr19_-_12078583 | 0.04 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr9_-_2945008 | 0.04 |
ENSDART00000183452
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr18_+_19006063 | 0.04 |
ENSDART00000135729
|
slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr4_+_17353714 | 0.04 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr10_+_6013076 | 0.04 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr8_-_19649617 | 0.04 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr9_+_10014514 | 0.04 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr10_+_16092671 | 0.04 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
| chr6_-_16406210 | 0.04 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr23_+_5465806 | 0.04 |
ENSDART00000149434
ENSDART00000148506 |
tulp1a
|
tubby like protein 1a |
| chr7_+_39011355 | 0.04 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr12_+_2660733 | 0.04 |
ENSDART00000152235
|
irbpl
|
interphotoreceptor retinoid-binding protein like |
| chr2_-_32551178 | 0.04 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr4_-_49133107 | 0.04 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr4_+_63484571 | 0.04 |
ENSDART00000169518
ENSDART00000168681 |
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr20_+_42537768 | 0.04 |
ENSDART00000134066
ENSDART00000153434 |
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr3_-_3366590 | 0.03 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr4_-_56083825 | 0.03 |
ENSDART00000157710
|
znf986
|
zinc finger protein 986 |
| chr19_-_2876321 | 0.03 |
ENSDART00000159253
|
exosc7
|
exosome component 7 |
| chr20_-_54924593 | 0.03 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr17_+_18031304 | 0.03 |
ENSDART00000127259
|
setd3
|
SET domain containing 3 |
| chr13_-_36844945 | 0.03 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr5_-_65081600 | 0.03 |
ENSDART00000160850
|
inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr23_+_2560005 | 0.03 |
ENSDART00000186906
|
GGT7
|
gamma-glutamyltransferase 7 |
| chr10_+_42898103 | 0.03 |
ENSDART00000015872
|
zcchc9
|
zinc finger, CCHC domain containing 9 |
| chr13_+_1015749 | 0.03 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr13_+_12528043 | 0.03 |
ENSDART00000057761
|
rrh
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr13_-_8815530 | 0.03 |
ENSDART00000122498
|
si:ch73-105b23.6
|
si:ch73-105b23.6 |
| chr7_-_30492261 | 0.03 |
ENSDART00000173954
|
adam10a
|
ADAM metallopeptidase domain 10a |
| chr23_+_12361899 | 0.03 |
ENSDART00000143728
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
Network of associatons between targets according to the STRING database.
First level regulatory network of crx+otx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.3 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.0 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.0 | 0.3 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 0.0 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.0 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.0 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.3 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.5 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.0 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |