Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
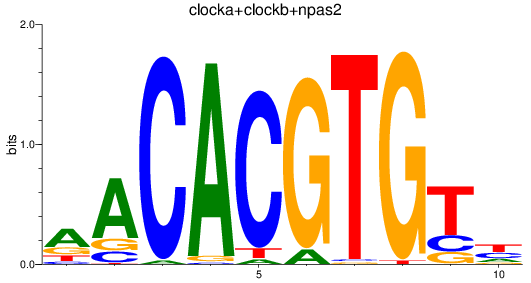
Results for clocka+clockb+npas2
Z-value: 2.09
Transcription factors associated with clocka+clockb+npas2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
clockb
|
ENSDARG00000003631 | clock circadian regulator b |
|
clocka
|
ENSDARG00000011703 | clock circadian regulator a |
|
npas2
|
ENSDARG00000016536 | neuronal PAS domain protein 2 |
|
npas2
|
ENSDARG00000116993 | neuronal PAS domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| npas2 | dr11_v1_chr5_+_22791686_22791686 | -0.97 | 7.5e-03 | Click! |
| clocka | dr11_v1_chr20_+_22130284_22130284 | -0.81 | 9.9e-02 | Click! |
| clockb | dr11_v1_chr1_+_19433004_19433004 | -0.69 | 2.0e-01 | Click! |
Activity profile of clocka+clockb+npas2 motif
Sorted Z-values of clocka+clockb+npas2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_43119698 | 2.16 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr10_+_22034477 | 1.72 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr14_+_22076596 | 1.59 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr24_-_12938922 | 1.36 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr19_-_47555956 | 1.30 |
ENSDART00000114549
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr2_-_42375275 | 1.25 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr8_+_26059677 | 1.21 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr5_+_38462121 | 1.18 |
ENSDART00000144425
|
gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr8_-_21372446 | 1.18 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr11_-_16975190 | 1.12 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr8_+_23213320 | 1.04 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr20_-_29483514 | 1.04 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr10_+_17026870 | 1.00 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr19_+_42983613 | 0.99 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr6_+_50451337 | 0.97 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr22_+_835728 | 0.91 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr6_-_8498908 | 0.89 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr12_+_19384615 | 0.89 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr22_-_23612854 | 0.87 |
ENSDART00000165885
|
cfhl5
|
complement factor H like 5 |
| chr21_-_20765338 | 0.83 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr21_+_11969603 | 0.82 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr3_+_18398876 | 0.81 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr7_-_32021853 | 0.78 |
ENSDART00000134521
|
kif18a
|
kinesin family member 18A |
| chr11_-_37995501 | 0.77 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr23_+_25292147 | 0.76 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr25_+_28555584 | 0.76 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr23_+_17417539 | 0.75 |
ENSDART00000182605
|
BX649300.2
|
|
| chr5_+_19337108 | 0.74 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr22_+_661711 | 0.74 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr1_-_52461322 | 0.73 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr20_-_49704915 | 0.72 |
ENSDART00000189232
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr18_+_15644559 | 0.72 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_-_32505109 | 0.72 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr5_-_1963498 | 0.72 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr5_-_8817458 | 0.72 |
ENSDART00000191098
|
fgf10b
|
fibroblast growth factor 10b |
| chr22_+_661505 | 0.72 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr3_+_1015867 | 0.72 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr18_-_19456269 | 0.71 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr2_+_37875789 | 0.71 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr17_+_38566717 | 0.70 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr21_+_43702016 | 0.69 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr22_+_21317597 | 0.69 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr5_-_29534748 | 0.69 |
ENSDART00000159587
|
AL831768.1
|
|
| chr17_-_49407091 | 0.69 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr9_+_34996245 | 0.68 |
ENSDART00000111437
|
jam2b
|
junctional adhesion molecule 2b |
| chr2_-_51630555 | 0.68 |
ENSDART00000171746
|
pigr
|
polymeric immunoglobulin receptor |
| chr24_-_42090635 | 0.67 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr19_-_24555935 | 0.67 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr20_+_10544100 | 0.67 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr22_-_17489040 | 0.67 |
ENSDART00000141286
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr11_-_29946927 | 0.66 |
ENSDART00000165182
|
zgc:113276
|
zgc:113276 |
| chr7_+_6652967 | 0.65 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr7_+_67467702 | 0.65 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr10_-_322769 | 0.65 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr6_-_52348562 | 0.64 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr11_+_29671661 | 0.64 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr11_-_29946640 | 0.64 |
ENSDART00000079175
|
zgc:113276
|
zgc:113276 |
| chr22_-_5323482 | 0.64 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr13_+_15701849 | 0.63 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr13_-_31370184 | 0.63 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr3_-_16760923 | 0.62 |
ENSDART00000055855
|
aspdh
|
aspartate dehydrogenase domain containing |
| chr3_-_3398383 | 0.62 |
ENSDART00000047865
|
si:dkey-46g23.2
|
si:dkey-46g23.2 |
| chr5_+_29714786 | 0.62 |
ENSDART00000148314
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr9_+_42270043 | 0.61 |
ENSDART00000137435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr7_-_38644560 | 0.60 |
ENSDART00000114934
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr14_+_22022441 | 0.60 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr6_-_49634787 | 0.60 |
ENSDART00000188538
|
BX323459.1
|
|
| chr3_-_34027178 | 0.60 |
ENSDART00000170201
ENSDART00000151408 |
ighv1-4
ighv14-1
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 14-1 |
| chr4_-_16824231 | 0.60 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr24_-_38644937 | 0.59 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr16_+_40954481 | 0.59 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr16_-_31919568 | 0.59 |
ENSDART00000027364
|
rbfox1l
|
RNA binding fox-1 homolog 1, like |
| chr9_+_23770666 | 0.58 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr20_+_46660339 | 0.58 |
ENSDART00000016530
|
adcy3b
|
adenylate cyclase 3b |
| chr13_-_42306348 | 0.58 |
ENSDART00000003706
|
kmo
|
kynurenine 3-monooxygenase |
| chr11_+_3959495 | 0.58 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr4_-_16824556 | 0.58 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr19_-_24555623 | 0.57 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr22_-_20695237 | 0.56 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr19_+_32166702 | 0.56 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr14_+_14836468 | 0.56 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr17_+_53156530 | 0.56 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr19_+_20201254 | 0.56 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr19_+_43123331 | 0.55 |
ENSDART00000187836
|
CABZ01101996.1
|
|
| chr5_-_32505276 | 0.55 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr17_-_51651631 | 0.55 |
ENSDART00000154699
|
ccr6b
|
chemokine (C-C motif) receptor 6b |
| chr11_-_28050559 | 0.54 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr15_+_20239141 | 0.54 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr16_-_48400639 | 0.53 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr3_+_16229911 | 0.53 |
ENSDART00000121728
|
rpl19
|
ribosomal protein L19 |
| chr7_-_38644287 | 0.53 |
ENSDART00000182307
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr12_+_13091842 | 0.52 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr16_+_33902006 | 0.52 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr23_-_29824146 | 0.52 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr5_+_29715040 | 0.52 |
ENSDART00000192563
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr10_-_15340362 | 0.52 |
ENSDART00000148119
ENSDART00000127277 ENSDART00000154037 ENSDART00000189109 |
pum3
|
pumilio RNA-binding family member 3 |
| chr16_-_13388821 | 0.52 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr2_+_25839650 | 0.52 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr25_-_30344117 | 0.52 |
ENSDART00000167077
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr5_+_72194444 | 0.52 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr14_+_23518110 | 0.51 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr6_+_29305190 | 0.51 |
ENSDART00000078647
|
si:ch211-201h21.5
|
si:ch211-201h21.5 |
| chr9_-_33328948 | 0.50 |
ENSDART00000006948
|
rpl8
|
ribosomal protein L8 |
| chr9_-_56231387 | 0.50 |
ENSDART00000149851
|
rpl31
|
ribosomal protein L31 |
| chr21_-_22724980 | 0.50 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr5_-_16475374 | 0.50 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr6_+_3717613 | 0.50 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr4_+_25607743 | 0.50 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr15_-_14038227 | 0.50 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr6_-_21189295 | 0.50 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr18_+_40993196 | 0.50 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr10_-_373575 | 0.50 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr11_+_12052791 | 0.49 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr15_-_17099560 | 0.49 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr21_+_26071874 | 0.49 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr19_-_42573219 | 0.49 |
ENSDART00000126021
ENSDART00000133695 ENSDART00000131558 |
zgc:103438
|
zgc:103438 |
| chr13_-_24260609 | 0.48 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr3_-_25377163 | 0.48 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr20_-_33675676 | 0.48 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr9_+_20781047 | 0.48 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr11_-_12800945 | 0.47 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr23_+_25291891 | 0.47 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr15_+_25635326 | 0.47 |
ENSDART00000135409
ENSDART00000162240 ENSDART00000052645 |
tsr1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr3_-_58798377 | 0.47 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr9_-_33329700 | 0.47 |
ENSDART00000147265
ENSDART00000140039 |
rpl8
|
ribosomal protein L8 |
| chr12_-_18898413 | 0.47 |
ENSDART00000181281
ENSDART00000121866 |
desi1b
|
desumoylating isopeptidase 1b |
| chr3_-_18805225 | 0.47 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr5_+_12836913 | 0.46 |
ENSDART00000023101
|
pes
|
pescadillo |
| chr6_-_57476465 | 0.46 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr5_-_2689753 | 0.46 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr1_-_26675969 | 0.45 |
ENSDART00000054184
|
trmo
|
tRNA methyltransferase O |
| chr3_+_1150348 | 0.44 |
ENSDART00000148524
|
nol12
|
nucleolar protein 12 |
| chr1_+_18550864 | 0.44 |
ENSDART00000142515
|
si:dkey-192k22.2
|
si:dkey-192k22.2 |
| chr3_-_42981739 | 0.44 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr11_-_44194132 | 0.44 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr23_+_16935494 | 0.44 |
ENSDART00000143120
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr20_+_25626479 | 0.44 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr1_-_51219965 | 0.44 |
ENSDART00000146612
|
esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr9_+_21358941 | 0.44 |
ENSDART00000147619
ENSDART00000059402 |
eef1akmt1
|
EEF1A lysine methyltransferase 1 |
| chr3_+_54581987 | 0.44 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr11_-_12801157 | 0.44 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr25_+_16356083 | 0.44 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr24_+_34069675 | 0.44 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr17_+_24821627 | 0.44 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr1_-_9195629 | 0.43 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr22_-_23591340 | 0.43 |
ENSDART00000167024
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr7_-_30779575 | 0.43 |
ENSDART00000004782
|
mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr8_+_17184602 | 0.43 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr3_-_58165254 | 0.42 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr17_-_45370200 | 0.42 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr2_+_5793908 | 0.41 |
ENSDART00000145219
|
slc35d1b
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1b |
| chr13_-_31025505 | 0.41 |
ENSDART00000137709
|
wdfy4
|
WDFY family member 4 |
| chr5_+_37744625 | 0.41 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr18_+_40993369 | 0.41 |
ENSDART00000141162
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr16_-_34195002 | 0.41 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr19_+_43579786 | 0.41 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr1_+_10297027 | 0.41 |
ENSDART00000152562
|
eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr24_-_20016817 | 0.40 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr21_-_41838284 | 0.40 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr11_+_2180072 | 0.40 |
ENSDART00000186854
ENSDART00000164839 |
hoxc11b
|
homeobox C11b |
| chr22_-_36690742 | 0.40 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr1_-_19764038 | 0.40 |
ENSDART00000005733
|
tma16
|
translation machinery associated 16 homolog |
| chr20_+_33875256 | 0.40 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr18_+_38191346 | 0.40 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr8_-_46486009 | 0.39 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr7_+_1505507 | 0.39 |
ENSDART00000161015
|
nop10
|
NOP10 ribonucleoprotein homolog (yeast) |
| chr3_-_58798815 | 0.39 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr22_+_29991834 | 0.39 |
ENSDART00000147728
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr20_+_38257766 | 0.39 |
ENSDART00000147485
ENSDART00000149160 |
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr7_+_32021982 | 0.39 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr5_+_13870340 | 0.39 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr14_+_30774515 | 0.39 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr2_+_25839940 | 0.39 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr2_+_45191049 | 0.39 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr6_-_18531349 | 0.39 |
ENSDART00000160693
ENSDART00000169780 |
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr5_-_24270989 | 0.38 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr14_+_6423973 | 0.38 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr8_+_44783424 | 0.38 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr12_+_2677303 | 0.38 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr1_-_26676391 | 0.38 |
ENSDART00000152492
|
trmo
|
tRNA methyltransferase O |
| chr15_-_34930727 | 0.38 |
ENSDART00000179723
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr13_+_13930263 | 0.38 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr5_+_27488975 | 0.38 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr22_+_9751117 | 0.38 |
ENSDART00000121827
|
BX664625.1
|
|
| chr10_+_17714866 | 0.38 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr3_+_22375596 | 0.38 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr12_-_21684197 | 0.37 |
ENSDART00000152999
ENSDART00000153109 ENSDART00000148698 |
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr3_-_56871330 | 0.37 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr5_-_33959868 | 0.37 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr13_-_293250 | 0.37 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr25_+_19733704 | 0.37 |
ENSDART00000145334
ENSDART00000138946 ENSDART00000138851 |
zgc:101783
|
zgc:101783 |
| chr9_+_12894578 | 0.37 |
ENSDART00000081270
|
si:ch211-167j6.5
|
si:ch211-167j6.5 |
| chr19_+_48359259 | 0.37 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr8_+_44358443 | 0.37 |
ENSDART00000189130
ENSDART00000189212 |
CU914776.2
|
|
| chr9_+_29985010 | 0.37 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr19_+_16015881 | 0.37 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr13_+_15702142 | 0.37 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr5_-_67499279 | 0.37 |
ENSDART00000128050
|
si:dkey-251i10.3
|
si:dkey-251i10.3 |
| chr12_+_29240124 | 0.37 |
ENSDART00000053761
ENSDART00000130172 |
bms1
|
BMS1 ribosome biogenesis factor |
| chr12_+_23892972 | 0.37 |
ENSDART00000152852
|
svila
|
supervillin a |
Network of associatons between targets according to the STRING database.
First level regulatory network of clocka+clockb+npas2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.4 | 1.3 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.4 | 2.3 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.3 | 1.4 | GO:0071548 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.3 | 0.9 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.4 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.7 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 0.7 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 0.5 | GO:1905133 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 1.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.5 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 1.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.6 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.1 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 1.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.6 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 0.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.8 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.2 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 2.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.4 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.3 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.8 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.6 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.3 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.2 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.2 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.4 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.3 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 1.2 | GO:0009749 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.6 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.6 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.2 | GO:0071051 | U4 snRNA 3'-end processing(GO:0034475) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 9.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 1.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.3 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.4 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.2 | GO:0019474 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.1 | GO:0071634 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 1.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.4 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.6 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.4 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.0 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.9 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.7 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 1.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 4.1 | GO:0043043 | peptide biosynthetic process(GO:0043043) |
| 0.0 | 0.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.2 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.3 | 1.4 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 0.8 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.2 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 2.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.5 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 2.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 4.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 10.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.4 | 1.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.3 | 0.9 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 1.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 1.8 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.2 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 1.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 0.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 0.8 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.2 | 0.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.5 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.5 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.5 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.4 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.2 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.7 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 3.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.6 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.9 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 1.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 2.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.6 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 6.4 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |