Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
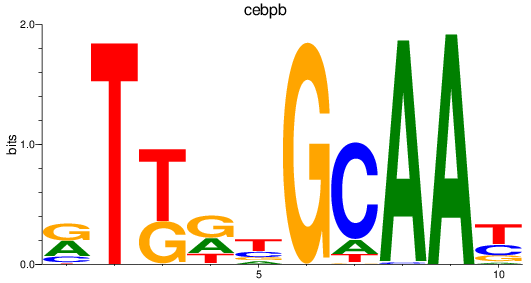
Results for cebpb
Z-value: 1.22
Transcription factors associated with cebpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpb
|
ENSDARG00000042725 | CCAAT enhancer binding protein beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpb | dr11_v1_chr8_-_28449782_28449782 | 0.53 | 3.6e-01 | Click! |
Activity profile of cebpb motif
Sorted Z-values of cebpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_23440632 | 1.96 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr5_-_63509581 | 1.73 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr14_+_21106444 | 1.33 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr14_+_16345003 | 1.32 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr14_+_21107032 | 1.26 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr9_-_32847642 | 1.11 |
ENSDART00000121506
|
hpx
|
hemopexin |
| chr8_-_50147948 | 1.08 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr6_+_21202639 | 1.03 |
ENSDART00000083126
|
cidec
|
cell death-inducing DFFA-like effector c |
| chr21_+_20901505 | 1.02 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr16_-_21785261 | 1.02 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr13_-_20540790 | 1.01 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr3_+_19299309 | 1.00 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr9_+_38292947 | 0.95 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr7_-_60831082 | 0.88 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr20_-_40755614 | 0.85 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr1_-_10071422 | 0.83 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr16_-_16701718 | 0.81 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr22_-_26175237 | 0.72 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr2_+_11029138 | 0.71 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr20_+_10538025 | 0.70 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr3_-_40275096 | 0.69 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr9_-_53062083 | 0.69 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr20_+_28266892 | 0.67 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr8_-_39739627 | 0.65 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr16_+_25245857 | 0.65 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr6_+_22068589 | 0.65 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr19_-_5699703 | 0.65 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr7_-_7845540 | 0.65 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr20_+_10544100 | 0.64 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr16_+_50289916 | 0.63 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr7_+_27691647 | 0.60 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr14_+_36885524 | 0.60 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr9_-_48736388 | 0.58 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr5_-_69948099 | 0.57 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr16_+_23984179 | 0.57 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr17_+_30450163 | 0.55 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr25_-_32311048 | 0.55 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr20_+_37794633 | 0.53 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr2_-_32643738 | 0.51 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr1_-_59126139 | 0.50 |
ENSDART00000156105
|
si:ch1073-110a20.7
|
si:ch1073-110a20.7 |
| chr22_-_26236188 | 0.49 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr7_+_12950507 | 0.48 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr19_+_5674907 | 0.48 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr10_+_38593645 | 0.48 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr5_-_43935119 | 0.48 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr7_-_4296771 | 0.47 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr5_+_26795773 | 0.46 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr15_-_1885247 | 0.46 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr9_-_1978090 | 0.45 |
ENSDART00000082344
|
hoxd11a
|
homeobox D11a |
| chr16_-_27749172 | 0.44 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr22_+_18469004 | 0.44 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr6_-_43449013 | 0.44 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr8_+_30699429 | 0.43 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr18_+_15644559 | 0.43 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr10_+_38610741 | 0.43 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr7_+_46019780 | 0.43 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr23_+_36095260 | 0.42 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr1_-_59104145 | 0.42 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr19_-_9662958 | 0.42 |
ENSDART00000041094
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr8_-_26961779 | 0.41 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr24_+_9881219 | 0.41 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr9_-_9989660 | 0.40 |
ENSDART00000081463
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr5_+_28830388 | 0.39 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr2_-_127945 | 0.38 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr4_-_19693978 | 0.36 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr14_+_48862987 | 0.36 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr17_-_15149192 | 0.36 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr12_+_31616412 | 0.35 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr22_-_17677947 | 0.35 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr5_-_43935460 | 0.35 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr17_+_10318071 | 0.35 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr10_+_4924388 | 0.35 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr1_-_59130695 | 0.35 |
ENSDART00000152560
|
FP015850.1
|
|
| chr6_-_13498745 | 0.34 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr24_-_38644937 | 0.34 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr5_+_8196264 | 0.33 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr10_+_17088261 | 0.33 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
| chr16_+_44298902 | 0.33 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr3_+_14641962 | 0.33 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr23_+_24272421 | 0.33 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr7_+_49862837 | 0.31 |
ENSDART00000174315
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr15_-_29348212 | 0.31 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr14_-_46238186 | 0.30 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr5_-_56412262 | 0.30 |
ENSDART00000083079
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr7_+_46020508 | 0.29 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr18_+_17493859 | 0.29 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr4_-_77432218 | 0.29 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr17_-_34952562 | 0.29 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr12_-_22238004 | 0.28 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr12_+_4920451 | 0.28 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr20_+_47491247 | 0.28 |
ENSDART00000113412
|
lin28b
|
lin-28 homolog B (C. elegans) |
| chr12_+_25775734 | 0.28 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr21_+_4509483 | 0.28 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr10_-_23099809 | 0.28 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr15_+_32711663 | 0.27 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr5_+_51079504 | 0.27 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr19_+_8606883 | 0.27 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr7_-_46019756 | 0.27 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr14_+_22022441 | 0.27 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr11_-_12198765 | 0.26 |
ENSDART00000104203
ENSDART00000128364 ENSDART00000166887 ENSDART00000041533 |
krt95
|
kertain 95 |
| chr25_+_31323978 | 0.26 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr16_+_20895904 | 0.26 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr3_+_22242269 | 0.26 |
ENSDART00000168970
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr8_+_25351863 | 0.25 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr25_-_27843066 | 0.25 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr3_-_57744323 | 0.25 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr13_+_41917606 | 0.25 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr1_-_59134522 | 0.24 |
ENSDART00000152764
|
si:ch1073-110a20.2
|
si:ch1073-110a20.2 |
| chr15_-_18232712 | 0.24 |
ENSDART00000081199
|
wu:fj20b03
|
wu:fj20b03 |
| chr7_+_11543999 | 0.24 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr6_+_53288018 | 0.24 |
ENSDART00000170319
|
cysltr3
|
cysteinyl leukotriene receptor 3 |
| chr12_-_8070969 | 0.24 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr4_+_9508505 | 0.23 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr14_+_10505464 | 0.23 |
ENSDART00000091224
|
p2ry10
|
P2Y receptor family member 10 |
| chr10_+_23099890 | 0.23 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr24_-_32582880 | 0.23 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr24_-_33284945 | 0.23 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr1_-_52222989 | 0.23 |
ENSDART00000010236
|
cnn1a
|
calponin 1, basic, smooth muscle, a |
| chr8_-_19280856 | 0.22 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr11_+_7214353 | 0.22 |
ENSDART00000156764
|
nwd1
|
NACHT and WD repeat domain containing 1 |
| chr22_-_17595310 | 0.22 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr8_-_28349859 | 0.22 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr1_-_59116617 | 0.22 |
ENSDART00000137471
ENSDART00000140490 |
MFAP4 (1 of many)
|
si:zfos-2330d3.7 |
| chr22_-_10487490 | 0.22 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr3_+_3641429 | 0.21 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr24_-_32582378 | 0.21 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr4_+_77973876 | 0.21 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr23_+_38957738 | 0.21 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr24_+_16983269 | 0.21 |
ENSDART00000139176
ENSDART00000023833 |
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr22_-_14361022 | 0.21 |
ENSDART00000140224
|
si:dkeyp-122a9.2
|
si:dkeyp-122a9.2 |
| chr5_-_32336613 | 0.21 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr19_-_15192840 | 0.21 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr3_+_3573696 | 0.21 |
ENSDART00000169680
|
CR589947.2
|
|
| chr1_+_8534698 | 0.21 |
ENSDART00000021504
|
smcr8b
|
Smith-Magenis syndrome chromosome region, candidate 8b |
| chr3_+_23703704 | 0.20 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr9_+_13682133 | 0.20 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr15_-_12258851 | 0.20 |
ENSDART00000180656
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr1_+_14253118 | 0.20 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr20_-_26531850 | 0.20 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr4_-_12725513 | 0.20 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr24_+_37406535 | 0.20 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr1_-_9231952 | 0.20 |
ENSDART00000166515
|
si:dkeyp-57d7.4
|
si:dkeyp-57d7.4 |
| chr20_-_52271262 | 0.20 |
ENSDART00000135463
|
rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr1_+_2260407 | 0.20 |
ENSDART00000058876
|
kpnb3
|
karyopherin (importin) beta 3 |
| chr12_-_5120175 | 0.20 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr1_-_59130383 | 0.20 |
ENSDART00000171552
|
FP015850.1
|
|
| chr21_+_17051478 | 0.19 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr8_+_20438884 | 0.19 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr20_-_26532167 | 0.19 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr6_+_8314451 | 0.19 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr17_+_3124129 | 0.19 |
ENSDART00000155323
|
zgc:136872
|
zgc:136872 |
| chr23_+_25305431 | 0.19 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr4_-_20181964 | 0.19 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr9_-_5318873 | 0.19 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr25_-_16581233 | 0.19 |
ENSDART00000155312
|
smc1b
|
structural maintenance of chromosomes 1B |
| chr7_+_15736230 | 0.18 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr18_-_18667416 | 0.18 |
ENSDART00000100401
ENSDART00000136544 |
aars
|
alanyl-tRNA synthetase |
| chr25_-_30429607 | 0.18 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr23_+_2669 | 0.18 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr23_+_30967686 | 0.18 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr13_+_33688474 | 0.18 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr25_-_17239008 | 0.18 |
ENSDART00000151965
ENSDART00000152106 ENSDART00000152107 |
ccnd2a
|
cyclin D2, a |
| chr17_+_443264 | 0.18 |
ENSDART00000159086
|
zgc:195050
|
zgc:195050 |
| chr2_-_40196547 | 0.18 |
ENSDART00000168098
|
ccl34a.3
|
chemokine (C-C motif) ligand 34a, duplicate 3 |
| chr1_-_59139848 | 0.17 |
ENSDART00000191863
|
si:ch1073-110a20.2
|
si:ch1073-110a20.2 |
| chr6_-_37422841 | 0.17 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr15_-_17960228 | 0.17 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr20_-_52271015 | 0.17 |
ENSDART00000074307
|
rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr12_+_46462090 | 0.17 |
ENSDART00000130748
|
BX005305.2
|
|
| chr7_+_16033923 | 0.17 |
ENSDART00000161669
ENSDART00000114062 |
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr4_+_8670662 | 0.17 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
| chr3_-_26204867 | 0.17 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr22_-_37686966 | 0.17 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr1_-_25177086 | 0.17 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr20_-_32045057 | 0.17 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr19_+_4990496 | 0.16 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr15_-_4314042 | 0.16 |
ENSDART00000173311
ENSDART00000170562 |
ECE2
|
si:ch211-117a13.2 |
| chr3_+_19216567 | 0.16 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr10_-_29733194 | 0.16 |
ENSDART00000149252
|
si:ch73-261i21.2
|
si:ch73-261i21.2 |
| chr5_-_20195350 | 0.16 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr12_+_16132612 | 0.16 |
ENSDART00000152550
|
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr12_+_46960579 | 0.16 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr7_+_65240227 | 0.15 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr8_-_46386024 | 0.15 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr12_-_10409961 | 0.15 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr4_+_71989418 | 0.15 |
ENSDART00000170996
|
parp11
|
poly(ADP-ribose) polymerase family member 11 |
| chr4_+_21866851 | 0.15 |
ENSDART00000177640
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr21_-_40174647 | 0.15 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr3_+_3992590 | 0.15 |
ENSDART00000180211
ENSDART00000054840 |
BX005421.2
|
|
| chr20_-_33675676 | 0.15 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr16_+_25259313 | 0.15 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr14_-_897874 | 0.15 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr6_+_8315050 | 0.15 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr23_+_11285662 | 0.14 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr10_+_21899753 | 0.14 |
ENSDART00000080155
|
hrh2b
|
histamine receptor H2b |
| chr23_+_38957472 | 0.14 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr6_+_39114345 | 0.14 |
ENSDART00000136835
|
gpr84
|
G protein-coupled receptor 84 |
| chr13_-_45155792 | 0.14 |
ENSDART00000163556
|
runx3
|
runt-related transcription factor 3 |
| chr22_+_15973122 | 0.13 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr10_+_15310811 | 0.13 |
ENSDART00000136239
|
kcnv2a
|
potassium channel, subfamily V, member 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 0.6 | GO:0009258 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 1.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.6 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 2.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 1.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.5 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 0.3 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.3 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 1.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 0.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.2 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.0 | 0.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.3 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 1.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.4 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.4 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) positive regulation of lipid transport(GO:0032370) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.6 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.1 | GO:0032602 | chemokine production(GO:0032602) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.2 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.4 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.0 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.9 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.1 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0055014 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.9 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.0 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.2 | 0.7 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.2 | 0.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.7 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.5 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 4.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 1.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |