Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
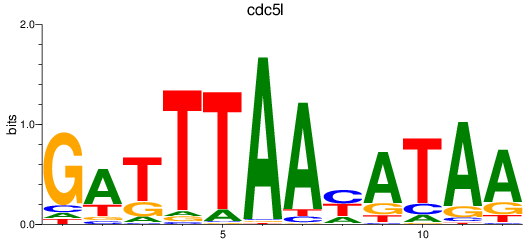
Results for cdc5l
Z-value: 1.94
Transcription factors associated with cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdc5l
|
ENSDARG00000043797 | CDC5 cell division cycle 5-like (S. pombe) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdc5l | dr11_v1_chr17_+_5061135_5061135 | -0.00 | 1.0e+00 | Click! |
Activity profile of cdc5l motif
Sorted Z-values of cdc5l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_33606739 | 2.05 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr19_-_31372896 | 2.04 |
ENSDART00000046609
|
scin
|
scinderin |
| chr16_-_25085327 | 1.99 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr1_-_45177373 | 1.37 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr18_+_7591381 | 1.34 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr6_-_49063085 | 1.26 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr22_+_7439186 | 1.23 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr10_+_2669405 | 1.19 |
ENSDART00000110202
|
ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr8_-_29719393 | 1.16 |
ENSDART00000077635
|
si:ch211-103n10.5
|
si:ch211-103n10.5 |
| chr12_+_46512881 | 1.12 |
ENSDART00000105454
|
CU855711.1
|
|
| chr12_+_5708400 | 1.10 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr15_-_12360409 | 1.03 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr23_+_5631381 | 1.03 |
ENSDART00000149143
|
pkp1a
|
plakophilin 1a |
| chr16_-_45230084 | 1.00 |
ENSDART00000184652
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr1_-_52447364 | 0.95 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr17_-_32426392 | 0.92 |
ENSDART00000148455
ENSDART00000149885 ENSDART00000179314 |
grhl1
|
grainyhead-like transcription factor 1 |
| chr21_+_20771082 | 0.89 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr1_-_10647307 | 0.89 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr22_-_10580194 | 0.89 |
ENSDART00000105848
|
SHISA4 (1 of many)
|
si:dkey-42i9.7 |
| chr23_-_44903048 | 0.88 |
ENSDART00000149103
|
fhdc5
|
FH2 domain containing 5 |
| chr3_-_61162750 | 0.88 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr24_-_31904924 | 0.88 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr4_-_11580948 | 0.86 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr19_-_3244360 | 0.84 |
ENSDART00000142222
|
zgc:158445
|
zgc:158445 |
| chr24_-_38131381 | 0.83 |
ENSDART00000105666
|
crp6
|
C-reactive protein 6 |
| chr17_+_5985933 | 0.83 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr7_+_24881680 | 0.83 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr5_-_55560937 | 0.82 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr19_+_2835240 | 0.81 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr3_-_32957702 | 0.79 |
ENSDART00000146586
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr24_+_27511734 | 0.79 |
ENSDART00000105771
|
cxcl32b.1
|
chemokine (C-X-C motif) ligand 32b, duplicate 1 |
| chr9_+_52398531 | 0.78 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr12_-_30549022 | 0.76 |
ENSDART00000102474
|
zgc:158404
|
zgc:158404 |
| chr16_-_12060770 | 0.76 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr16_+_55059026 | 0.75 |
ENSDART00000109391
|
LO017815.1
|
Danio rerio nuclear receptor coactivator 7-like (LOC792958), mRNA. |
| chr2_-_19234329 | 0.73 |
ENSDART00000161106
ENSDART00000160060 ENSDART00000174552 |
cdc20
|
cell division cycle 20 homolog |
| chr12_+_46462090 | 0.73 |
ENSDART00000130748
|
BX005305.2
|
|
| chr13_-_37620091 | 0.72 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr1_-_38171648 | 0.69 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr4_-_17812131 | 0.69 |
ENSDART00000025731
|
spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr1_-_10647484 | 0.68 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr16_+_29509133 | 0.67 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr16_+_23947196 | 0.67 |
ENSDART00000103190
ENSDART00000132961 ENSDART00000147690 ENSDART00000142168 |
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr2_-_8648440 | 0.66 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr25_+_25202213 | 0.66 |
ENSDART00000150744
|
ccl39a.10
|
chemokine (C-C motif) ligand 39, duplicate 10 |
| chr2_+_40181633 | 0.66 |
ENSDART00000185494
|
FP236331.1
|
|
| chr16_-_26140768 | 0.65 |
ENSDART00000143960
|
cd79a
|
CD79a molecule, immunoglobulin-associated alpha |
| chr4_+_1461079 | 0.64 |
ENSDART00000082383
|
trim35-14
|
tripartite motif containing 35-14 |
| chr2_+_10134345 | 0.63 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr17_-_36529449 | 0.62 |
ENSDART00000187252
|
colec11
|
collectin sub-family member 11 |
| chr23_+_28381260 | 0.62 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr9_-_9415000 | 0.62 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr6_-_609880 | 0.62 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr6_+_56141852 | 0.61 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr15_+_28410664 | 0.61 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr15_-_47929455 | 0.61 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr15_-_34892664 | 0.60 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr16_+_35916371 | 0.60 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr8_+_47099033 | 0.60 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr3_+_62161184 | 0.60 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr21_+_28749720 | 0.59 |
ENSDART00000145178
|
zgc:100829
|
zgc:100829 |
| chr3_-_7990516 | 0.59 |
ENSDART00000167877
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr16_-_12060488 | 0.59 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr15_+_28096152 | 0.58 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr25_-_22191983 | 0.58 |
ENSDART00000191181
|
pkp3a
|
plakophilin 3a |
| chr8_+_39674707 | 0.57 |
ENSDART00000126301
ENSDART00000040330 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr22_+_1911269 | 0.57 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr1_-_18848955 | 0.57 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr24_+_12945803 | 0.56 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr18_-_46354269 | 0.56 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr15_+_934660 | 0.55 |
ENSDART00000154248
|
si:dkey-77f5.10
|
si:dkey-77f5.10 |
| chr23_+_32011768 | 0.55 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr1_+_9966384 | 0.55 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr3_-_26921475 | 0.54 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr6_-_436658 | 0.53 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr21_-_17296010 | 0.53 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr5_+_45918680 | 0.53 |
ENSDART00000036242
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr5_-_62317496 | 0.52 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr18_+_13203831 | 0.52 |
ENSDART00000032151
|
cotl1
|
coactosin-like F-actin binding protein 1 |
| chr21_-_5205617 | 0.52 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr11_-_10659195 | 0.52 |
ENSDART00000115255
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr6_+_40992409 | 0.52 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr16_-_32208728 | 0.52 |
ENSDART00000023995
|
calhm5.2
|
calcium homeostasis modulator family member 5, tandem duplicate 2 |
| chr15_-_41689981 | 0.52 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr5_-_61652254 | 0.52 |
ENSDART00000097364
|
drl
|
draculin |
| chr13_-_45155792 | 0.52 |
ENSDART00000163556
|
runx3
|
runt-related transcription factor 3 |
| chr7_-_8315179 | 0.51 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr3_-_24681404 | 0.51 |
ENSDART00000161612
|
BX569789.1
|
|
| chr18_+_36631923 | 0.51 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr16_-_11859309 | 0.51 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr14_+_15622817 | 0.51 |
ENSDART00000158624
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr2_+_50967947 | 0.51 |
ENSDART00000162288
|
si:ch211-249o11.5
|
si:ch211-249o11.5 |
| chr3_-_54846444 | 0.51 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr10_+_8656417 | 0.50 |
ENSDART00000123131
|
pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr1_-_43915423 | 0.50 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr19_-_13933204 | 0.49 |
ENSDART00000158059
|
trnau1apa
|
tRNA selenocysteine 1 associated protein 1a |
| chr8_-_16725573 | 0.49 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr9_+_44994214 | 0.48 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr17_+_6563307 | 0.48 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr18_-_29925717 | 0.47 |
ENSDART00000099281
|
mhc2dbb
|
major histocompatibility complex class II DBB gene |
| chr11_+_37201483 | 0.47 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr21_-_28439596 | 0.47 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr5_+_21181047 | 0.47 |
ENSDART00000088506
|
arhgap25
|
Rho GTPase activating protein 25 |
| chr17_-_36529016 | 0.47 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr13_-_42757565 | 0.46 |
ENSDART00000161950
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr15_-_17813680 | 0.46 |
ENSDART00000158556
|
CT573342.2
|
|
| chr24_-_3426620 | 0.46 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr3_-_11203689 | 0.46 |
ENSDART00000166328
|
BX323882.1
|
|
| chr20_+_43691208 | 0.46 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr1_-_7570181 | 0.46 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr16_+_23398369 | 0.46 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr25_+_5249513 | 0.46 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr20_+_23238833 | 0.46 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr19_-_27588842 | 0.45 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr7_-_16598212 | 0.45 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr5_-_37117778 | 0.45 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr18_-_48296793 | 0.45 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr12_+_20641102 | 0.44 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr21_+_11560153 | 0.44 |
ENSDART00000065842
|
cd8a
|
CD8a molecule |
| chr3_+_22375596 | 0.44 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr22_-_22301672 | 0.44 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr4_-_38308760 | 0.43 |
ENSDART00000190780
|
si:ch211-229l10.4
|
si:ch211-229l10.4 |
| chr8_-_14554785 | 0.43 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr16_-_25741225 | 0.43 |
ENSDART00000130641
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr21_-_43457554 | 0.43 |
ENSDART00000085039
|
stk26
|
serine/threonine protein kinase 26 |
| chr2_+_31671545 | 0.43 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr16_-_46567344 | 0.42 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr4_-_11064073 | 0.42 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr19_+_7636941 | 0.42 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr11_+_14284866 | 0.42 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr4_-_76370630 | 0.42 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr4_+_5205265 | 0.41 |
ENSDART00000067382
|
galnt8b.1
|
polypeptide N-acetylgalactosaminyltransferase 8b, tandem duplicate 1 |
| chr22_+_30631072 | 0.41 |
ENSDART00000059970
|
zmp:0000000606
|
zmp:0000000606 |
| chr25_-_3217115 | 0.41 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr22_+_7742211 | 0.41 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr10_-_35153388 | 0.41 |
ENSDART00000188132
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr5_-_61624693 | 0.40 |
ENSDART00000141323
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr20_+_53522059 | 0.40 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr19_+_7045033 | 0.40 |
ENSDART00000146579
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr20_+_51104367 | 0.40 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr21_+_15295685 | 0.40 |
ENSDART00000135603
|
BX901878.1
|
|
| chr1_+_57187794 | 0.40 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr2_+_24507597 | 0.39 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr6_-_49526510 | 0.39 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr21_-_31226045 | 0.39 |
ENSDART00000011572
|
zgc:152891
|
zgc:152891 |
| chr2_+_45552901 | 0.39 |
ENSDART00000131701
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr4_-_13614797 | 0.39 |
ENSDART00000138366
ENSDART00000165212 |
irf5
|
interferon regulatory factor 5 |
| chr16_-_33104944 | 0.39 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr1_-_51734524 | 0.39 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr22_-_22301929 | 0.39 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr1_-_11104805 | 0.39 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr17_+_16090436 | 0.39 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr19_-_10551394 | 0.38 |
ENSDART00000186815
ENSDART00000181041 |
lim2.4
|
lens intrinsic membrane protein 2.4 |
| chr15_-_29573267 | 0.38 |
ENSDART00000099947
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr14_+_9456675 | 0.38 |
ENSDART00000134935
|
rell2
|
RELT-like 2 |
| chr1_-_53880639 | 0.38 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr9_+_38645136 | 0.38 |
ENSDART00000135505
|
slc12a8
|
solute carrier family 12, member 8 |
| chr25_+_35051656 | 0.38 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr2_-_10877228 | 0.37 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr2_-_5840494 | 0.37 |
ENSDART00000154877
|
si:ch211-273k1.5
|
si:ch211-273k1.5 |
| chr7_-_20464468 | 0.37 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr18_+_31073265 | 0.37 |
ENSDART00000023539
|
cyba
|
cytochrome b-245, alpha polypeptide |
| chr11_+_35050253 | 0.37 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr17_-_5424175 | 0.37 |
ENSDART00000171063
ENSDART00000188102 |
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr10_+_10313024 | 0.37 |
ENSDART00000142895
ENSDART00000129952 |
urm1
|
ubiquitin related modifier 1 |
| chr1_-_49505449 | 0.37 |
ENSDART00000187294
ENSDART00000132171 |
si:dkeyp-80c12.5
|
si:dkeyp-80c12.5 |
| chr2_-_36604589 | 0.37 |
ENSDART00000167352
|
tradv41.0
|
T-cell receptor alpha/delta variable 41.0 |
| chr17_-_15189397 | 0.36 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr15_+_37986069 | 0.36 |
ENSDART00000156984
|
si:dkey-238d18.8
|
si:dkey-238d18.8 |
| chr15_-_5815006 | 0.36 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr14_-_34633960 | 0.36 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr17_-_31695217 | 0.36 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr25_-_3228025 | 0.36 |
ENSDART00000165924
|
cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr20_+_19006703 | 0.36 |
ENSDART00000128435
|
pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr8_-_45756434 | 0.36 |
ENSDART00000179658
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr1_-_58064738 | 0.36 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr21_+_45816030 | 0.36 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr1_-_55008882 | 0.35 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr25_-_15040369 | 0.35 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr7_+_25053331 | 0.35 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr10_+_24627683 | 0.35 |
ENSDART00000112652
|
slc46a3
|
solute carrier family 46, member 3 |
| chr6_+_60125033 | 0.35 |
ENSDART00000148557
ENSDART00000008224 |
aurka
|
aurora kinase A |
| chr20_-_26491567 | 0.35 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr10_+_13209580 | 0.35 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr20_-_30377221 | 0.35 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr6_+_40992883 | 0.34 |
ENSDART00000076061
|
tgfa
|
transforming growth factor, alpha |
| chr15_-_41689684 | 0.34 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr10_-_21362320 | 0.34 |
ENSDART00000189789
|
avd
|
avidin |
| chr15_+_6652396 | 0.34 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr8_-_13010011 | 0.34 |
ENSDART00000140969
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr3_-_16537441 | 0.34 |
ENSDART00000080749
ENSDART00000133824 |
eps8l1
|
eps8-like1 |
| chr3_-_21382491 | 0.34 |
ENSDART00000016562
|
itga2b
|
integrin, alpha 2b |
| chr1_+_25178106 | 0.34 |
ENSDART00000054265
ENSDART00000141648 |
mnd1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae) |
| chr8_-_20245892 | 0.34 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr1_-_12161148 | 0.33 |
ENSDART00000166429
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr8_-_46486009 | 0.33 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr2_-_30182353 | 0.33 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr11_-_16152400 | 0.33 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr6_+_23712911 | 0.33 |
ENSDART00000167795
|
zgc:158654
|
zgc:158654 |
| chr2_-_47620806 | 0.33 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdc5l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 1.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 0.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.2 | 0.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.5 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.5 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.3 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 1.1 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.1 | 2.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.4 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.1 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.2 | GO:0045601 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.6 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.5 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.2 | GO:0016109 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.6 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.3 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.6 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.5 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.9 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.6 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.8 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.0 | 0.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.3 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.0 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.7 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0036473 | cell death in response to oxidative stress(GO:0036473) negative regulation of cellular response to oxidative stress(GO:1900408) negative regulation of response to oxidative stress(GO:1902883) regulation of oxidative stress-induced cell death(GO:1903201) negative regulation of oxidative stress-induced cell death(GO:1903202) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 1.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.3 | GO:0042744 | oxygen transport(GO:0015671) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.3 | GO:0009749 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.1 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.6 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.3 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.6 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0023058 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.0 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.9 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 2.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.4 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.3 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.6 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.0 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 1.6 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.0 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 0.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 0.9 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.7 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.4 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.3 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.2 | GO:0032356 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.2 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.2 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 2.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.6 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0008144 | drug binding(GO:0008144) cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.5 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |