Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
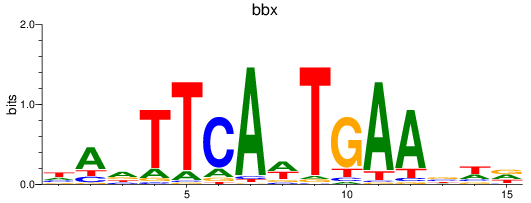
Results for bbx
Z-value: 0.86
Transcription factors associated with bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bbx
|
ENSDARG00000012699 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bbx | dr11_v1_chr10_-_28477023_28477023 | 0.68 | 2.1e-01 | Click! |
Activity profile of bbx motif
Sorted Z-values of bbx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_22648007 | 0.78 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr18_+_39060687 | 0.57 |
ENSDART00000098729
ENSDART00000136367 |
zgc:171509
|
zgc:171509 |
| chr21_-_22647695 | 0.47 |
ENSDART00000187553
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr16_+_23403602 | 0.44 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr21_-_22678195 | 0.41 |
ENSDART00000171231
|
gig2g
|
grass carp reovirus (GCRV)-induced gene 2g |
| chr7_+_72003301 | 0.40 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr8_-_36469117 | 0.39 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr16_-_40373836 | 0.32 |
ENSDART00000134498
|
si:dkey-242e21.3
|
si:dkey-242e21.3 |
| chr3_+_4113551 | 0.31 |
ENSDART00000192309
|
LO018551.1
|
|
| chr23_+_19670085 | 0.29 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr8_-_36327328 | 0.28 |
ENSDART00000183333
|
zgc:103700
|
zgc:103700 |
| chr11_+_2416064 | 0.27 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr19_-_40198478 | 0.27 |
ENSDART00000191736
|
grn2
|
granulin 2 |
| chr17_+_24718272 | 0.25 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr6_-_54816567 | 0.24 |
ENSDART00000150079
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr2_-_32237916 | 0.24 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr11_-_27501027 | 0.22 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr8_+_52701811 | 0.22 |
ENSDART00000179692
|
bcl2l16
|
BCL2 like 16 |
| chr6_+_36839509 | 0.22 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr19_+_43341424 | 0.22 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr16_-_47427016 | 0.21 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr8_-_14091886 | 0.21 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr7_-_54320088 | 0.20 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr20_-_25626693 | 0.20 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr16_-_45235947 | 0.20 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr5_-_42883761 | 0.19 |
ENSDART00000167374
|
BX323596.2
|
|
| chr6_-_19042294 | 0.19 |
ENSDART00000159461
|
si:rp71-81e14.2
|
si:rp71-81e14.2 |
| chr8_+_20951590 | 0.19 |
ENSDART00000138728
|
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr20_-_39735952 | 0.19 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr20_-_25626198 | 0.18 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr20_-_25626428 | 0.18 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr19_+_40856807 | 0.17 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr16_+_41570653 | 0.17 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr10_-_42751641 | 0.16 |
ENSDART00000182734
ENSDART00000113926 |
zgc:100918
|
zgc:100918 |
| chr5_+_36650096 | 0.16 |
ENSDART00000111414
|
alkbh6
|
alkB homolog 6 |
| chr13_-_7233811 | 0.16 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr5_+_69733096 | 0.16 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr1_+_55535827 | 0.16 |
ENSDART00000152784
|
adgre16
|
adhesion G protein-coupled receptor E16 |
| chr16_+_32014552 | 0.15 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr12_-_33770299 | 0.15 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr23_-_18381361 | 0.15 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr9_-_30555725 | 0.15 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr2_-_22659450 | 0.15 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr6_+_37301341 | 0.14 |
ENSDART00000104180
|
zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr12_+_27156943 | 0.14 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr10_+_23511816 | 0.14 |
ENSDART00000127255
|
zmp:0000000924
|
zmp:0000000924 |
| chr1_+_55643198 | 0.14 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr23_+_35504824 | 0.13 |
ENSDART00000082647
ENSDART00000159218 |
acp1
|
acid phosphatase 1 |
| chr3_+_13862753 | 0.13 |
ENSDART00000168315
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr14_-_4177311 | 0.13 |
ENSDART00000128129
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr21_+_21263988 | 0.13 |
ENSDART00000089651
ENSDART00000108978 |
ccdc61
|
coiled-coil domain containing 61 |
| chr1_+_2431956 | 0.12 |
ENSDART00000183832
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr23_+_3721042 | 0.12 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr8_-_40205712 | 0.11 |
ENSDART00000158927
|
anapc5
|
anaphase promoting complex subunit 5 |
| chr15_-_31109760 | 0.11 |
ENSDART00000154254
|
ksr1b
|
kinase suppressor of ras 1b |
| chr18_+_6857071 | 0.11 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr16_-_13622794 | 0.11 |
ENSDART00000146953
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr13_-_23095006 | 0.11 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr10_-_26218354 | 0.11 |
ENSDART00000180764
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr1_+_55563532 | 0.10 |
ENSDART00000152549
|
adgre15
|
adhesion G protein-coupled receptor E15 |
| chr7_-_50395059 | 0.10 |
ENSDART00000191150
|
vps33b
|
vacuolar protein sorting 33B |
| chr23_+_22656477 | 0.10 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr13_+_23095228 | 0.10 |
ENSDART00000189068
ENSDART00000188624 |
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr8_+_23034718 | 0.10 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr1_+_55583116 | 0.09 |
ENSDART00000152163
|
adgre19
|
adhesion G protein-coupled receptor E19 |
| chr22_+_9939901 | 0.09 |
ENSDART00000169777
ENSDART00000081420 |
zgc:171686
|
zgc:171686 |
| chr16_+_25535993 | 0.09 |
ENSDART00000077436
|
mylipb
|
myosin regulatory light chain interacting protein b |
| chr15_-_15983428 | 0.09 |
ENSDART00000115129
|
synrg
|
synergin, gamma |
| chr20_+_25626479 | 0.09 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr22_+_35930526 | 0.09 |
ENSDART00000169242
|
LO016987.1
|
|
| chr15_-_15983183 | 0.08 |
ENSDART00000154841
|
synrg
|
synergin, gamma |
| chr5_+_37744625 | 0.08 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr24_-_9002038 | 0.08 |
ENSDART00000066783
ENSDART00000150185 |
mppe1
|
metallophosphoesterase 1 |
| chr7_-_50395395 | 0.08 |
ENSDART00000065868
|
vps33b
|
vacuolar protein sorting 33B |
| chr7_+_6941583 | 0.08 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr15_-_4596623 | 0.08 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr13_-_50565338 | 0.07 |
ENSDART00000062684
|
blnk
|
B cell linker |
| chr16_+_16978424 | 0.07 |
ENSDART00000143128
|
rpl18
|
ribosomal protein L18 |
| chr22_+_19230434 | 0.07 |
ENSDART00000132929
|
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr7_+_50395856 | 0.07 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr14_-_30808174 | 0.07 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr13_+_18371208 | 0.07 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr23_+_2703044 | 0.07 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr18_+_26899316 | 0.07 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr12_+_4222854 | 0.07 |
ENSDART00000144881
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr20_-_20248408 | 0.06 |
ENSDART00000183234
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr6_-_35052388 | 0.06 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr3_-_37759969 | 0.06 |
ENSDART00000151105
ENSDART00000151208 |
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr17_-_24439672 | 0.06 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr16_+_16977786 | 0.06 |
ENSDART00000043173
ENSDART00000132150 |
rpl18
|
ribosomal protein L18 |
| chr23_-_24343363 | 0.06 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr7_+_11459235 | 0.05 |
ENSDART00000159611
|
il16
|
interleukin 16 |
| chr3_-_34816893 | 0.05 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr17_+_18117976 | 0.05 |
ENSDART00000123745
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr4_+_65061448 | 0.05 |
ENSDART00000172380
|
si:dkey-14o6.2
|
si:dkey-14o6.2 |
| chr1_+_45121393 | 0.05 |
ENSDART00000142702
|
muc13a
|
mucin 13a, cell surface associated |
| chr4_+_5215950 | 0.05 |
ENSDART00000140613
|
galnt8b.2
|
polypeptide N-acetylgalactosaminyltransferase 8b, tandem duplicate 2 |
| chr23_+_40139765 | 0.05 |
ENSDART00000185376
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr7_+_36898850 | 0.05 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr12_+_23812530 | 0.05 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr21_-_25416391 | 0.05 |
ENSDART00000114081
|
sms
|
spermine synthase |
| chr17_-_14523722 | 0.04 |
ENSDART00000024726
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr17_+_8799661 | 0.04 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr16_-_24195252 | 0.04 |
ENSDART00000136205
ENSDART00000048599 ENSDART00000161547 |
rps19
|
ribosomal protein S19 |
| chr1_+_23784905 | 0.04 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr4_+_34121902 | 0.04 |
ENSDART00000170225
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr7_-_50604626 | 0.04 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr8_-_36370552 | 0.04 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr6_-_39344259 | 0.04 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr13_+_31205439 | 0.04 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr4_+_57194439 | 0.04 |
ENSDART00000075265
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr17_+_8799451 | 0.04 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr6_+_6780873 | 0.03 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr9_+_40546677 | 0.03 |
ENSDART00000132911
|
vwc2l
|
von Willebrand factor C domain containing protein 2-like |
| chr5_+_61556172 | 0.03 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr12_-_17686404 | 0.03 |
ENSDART00000079065
|
ccz1
|
CCZ1 homolog, vacuolar protein trafficking and biogenesis associated |
| chr8_+_21114338 | 0.03 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| chr25_-_12730260 | 0.03 |
ENSDART00000171834
|
klhdc4
|
kelch domain containing 4 |
| chr6_+_14980761 | 0.03 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr5_-_30978381 | 0.03 |
ENSDART00000127787
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr19_-_3773905 | 0.03 |
ENSDART00000168433
|
btr20
|
bloodthirsty-related gene family, member 20 |
| chr19_+_10860485 | 0.03 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
| chr23_+_17512037 | 0.03 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr1_+_19433004 | 0.02 |
ENSDART00000133959
|
clockb
|
clock circadian regulator b |
| chr24_+_41989108 | 0.02 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr10_-_43771447 | 0.02 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr11_-_12104917 | 0.02 |
ENSDART00000136108
|
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr18_-_17724295 | 0.02 |
ENSDART00000121553
|
fam192a
|
family with sequence similarity 192, member A |
| chr4_+_70342815 | 0.02 |
ENSDART00000126434
|
si:ch211-76m11.3
|
si:ch211-76m11.3 |
| chr1_+_55662491 | 0.02 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr1_-_7673376 | 0.02 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr4_-_36032930 | 0.02 |
ENSDART00000191414
|
zgc:174180
|
zgc:174180 |
| chr4_+_75577480 | 0.02 |
ENSDART00000188196
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr24_-_28229618 | 0.01 |
ENSDART00000145290
|
bcl2a
|
BCL2, apoptosis regulator a |
| chr16_-_27566552 | 0.01 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr15_-_35212462 | 0.01 |
ENSDART00000043960
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr15_+_44201056 | 0.01 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr3_+_32553714 | 0.01 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr25_-_16076257 | 0.01 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr24_-_36727922 | 0.01 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr9_-_15424639 | 0.01 |
ENSDART00000124346
|
fn1a
|
fibronectin 1a |
| chr13_-_24669258 | 0.01 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr2_+_54804385 | 0.01 |
ENSDART00000172309
|
twsg1a
|
twisted gastrulation BMP signaling modulator 1a |
| chr17_+_21546993 | 0.01 |
ENSDART00000182387
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr25_+_16646113 | 0.01 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr4_+_76442674 | 0.01 |
ENSDART00000164825
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr1_+_52481332 | 0.00 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr16_-_41465542 | 0.00 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr17_+_44697604 | 0.00 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr13_+_18507592 | 0.00 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr23_+_579893 | 0.00 |
ENSDART00000189098
|
BX323461.1
|
|
| chr17_-_8656155 | 0.00 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr4_-_36033377 | 0.00 |
ENSDART00000164348
ENSDART00000187058 |
zgc:174180
|
zgc:174180 |
| chr4_-_42397126 | 0.00 |
ENSDART00000162437
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr12_-_31484677 | 0.00 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr4_+_47636303 | 0.00 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr4_-_67969695 | 0.00 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr8_-_33154677 | 0.00 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bbx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.0 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.0 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |