Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
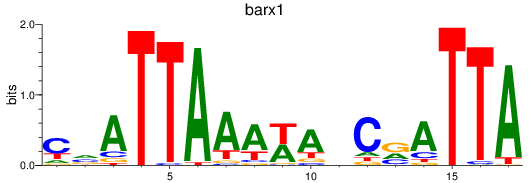
Results for barx1
Z-value: 1.32
Transcription factors associated with barx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barx1
|
ENSDARG00000007407 | BARX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barx1 | dr11_v1_chr11_+_27543093_27543093 | 0.16 | 8.0e-01 | Click! |
Activity profile of barx1 motif
Sorted Z-values of barx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_35019693 | 0.88 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr25_+_31276842 | 0.87 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr8_-_18537866 | 0.76 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr11_-_17981421 | 0.74 |
ENSDART00000005999
ENSDART00000104046 |
twf2b
|
twinfilin actin-binding protein 2b |
| chr22_-_31060579 | 0.73 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr5_+_2815021 | 0.72 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr23_+_409578 | 0.68 |
ENSDART00000008177
|
slc25a55b
|
solute carrier family 25, member 55b |
| chr3_+_59851537 | 0.66 |
ENSDART00000180997
|
CU693479.1
|
|
| chr14_-_970853 | 0.65 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr14_+_15622817 | 0.62 |
ENSDART00000158624
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr23_+_4689626 | 0.60 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr8_-_31062811 | 0.58 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr14_+_15430991 | 0.55 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr7_+_4474880 | 0.54 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr22_-_15587360 | 0.53 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr10_+_1396940 | 0.53 |
ENSDART00000150096
|
gdnfa
|
glial cell derived neurotrophic factor a |
| chr3_+_48206469 | 0.52 |
ENSDART00000161251
|
si:ch211-207b24.4
|
si:ch211-207b24.4 |
| chr16_-_25608453 | 0.51 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr16_-_38118003 | 0.50 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr4_+_2252123 | 0.49 |
ENSDART00000163996
ENSDART00000066491 |
glipr1a
|
GLI pathogenesis-related 1a |
| chr7_+_20471315 | 0.48 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr20_+_15015557 | 0.48 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr22_+_10651726 | 0.47 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr21_+_25765734 | 0.47 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr1_-_17735861 | 0.47 |
ENSDART00000018238
|
pdlim3a
|
PDZ and LIM domain 3a |
| chr13_-_4223955 | 0.46 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr7_-_16562200 | 0.46 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr24_+_21622373 | 0.45 |
ENSDART00000183611
|
rpl21
|
ribosomal protein L21 |
| chr22_+_12770877 | 0.45 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr19_-_40199081 | 0.45 |
ENSDART00000051970
ENSDART00000151079 |
grn2
|
granulin 2 |
| chr13_+_7442023 | 0.45 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr25_+_13620555 | 0.44 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr16_+_13965923 | 0.44 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr20_+_26556372 | 0.43 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr9_-_42484444 | 0.43 |
ENSDART00000048320
ENSDART00000047653 |
tfpia
|
tissue factor pathway inhibitor a |
| chr21_-_1635268 | 0.43 |
ENSDART00000151258
|
zgc:152948
|
zgc:152948 |
| chr4_+_76575585 | 0.42 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr18_+_5490668 | 0.42 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr12_-_26423439 | 0.41 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr3_+_24640946 | 0.41 |
ENSDART00000123994
ENSDART00000126230 |
zgc:113411
|
zgc:113411 |
| chr18_+_26719787 | 0.40 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr22_+_1170294 | 0.40 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr19_+_20778011 | 0.40 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr23_+_20422661 | 0.40 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr25_-_22187397 | 0.39 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr16_-_41762983 | 0.39 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr21_-_19316985 | 0.39 |
ENSDART00000141596
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr25_+_20089986 | 0.38 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr15_+_46313082 | 0.38 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr7_-_54320088 | 0.37 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr23_+_16935494 | 0.37 |
ENSDART00000143120
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr11_-_22303678 | 0.37 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr17_-_20143946 | 0.37 |
ENSDART00000138911
|
actn2b
|
actinin, alpha 2b |
| chr15_-_42206890 | 0.37 |
ENSDART00000015843
|
pax3b
|
paired box 3b |
| chr21_-_5881344 | 0.36 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr2_+_36109002 | 0.36 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr20_-_33976305 | 0.36 |
ENSDART00000111022
|
sele
|
selectin E |
| chr19_-_31372896 | 0.36 |
ENSDART00000046609
|
scin
|
scinderin |
| chr3_+_26081343 | 0.35 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr22_-_6499055 | 0.35 |
ENSDART00000142235
ENSDART00000183588 |
si:dkey-19a16.7
|
si:dkey-19a16.7 |
| chr6_+_39290777 | 0.35 |
ENSDART00000155700
|
si:dkey-195m11.11
|
si:dkey-195m11.11 |
| chr3_-_18030938 | 0.35 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr13_-_37615029 | 0.35 |
ENSDART00000111199
|
si:dkey-188i13.6
|
si:dkey-188i13.6 |
| chr1_+_58922027 | 0.35 |
ENSDART00000159479
|
trip10b
|
thyroid hormone receptor interactor 10b |
| chr10_+_33382858 | 0.35 |
ENSDART00000063662
|
mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr6_-_15096556 | 0.35 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr23_-_5719453 | 0.34 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr4_-_17785120 | 0.34 |
ENSDART00000024775
|
mybpc1
|
myosin binding protein C, slow type |
| chr9_+_6587364 | 0.34 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr17_+_25849332 | 0.34 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr5_+_32206378 | 0.34 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr10_+_38610741 | 0.34 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr16_-_27566552 | 0.33 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr11_+_42474694 | 0.33 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr22_-_36876133 | 0.33 |
ENSDART00000147006
|
kng1
|
kininogen 1 |
| chr11_+_43431289 | 0.33 |
ENSDART00000192526
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr6_-_49873020 | 0.33 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr24_+_28614576 | 0.32 |
ENSDART00000184941
|
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr20_+_38276690 | 0.32 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr20_+_38279523 | 0.32 |
ENSDART00000061311
|
ccl38a.5
|
chemokine (C-C motif) ligand 38, duplicate 5 |
| chr24_+_35387517 | 0.32 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr15_+_32711663 | 0.32 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr2_-_32237916 | 0.32 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr21_+_19525141 | 0.32 |
ENSDART00000058489
|
gzma
|
granzyme A |
| chr12_-_30043809 | 0.32 |
ENSDART00000150220
ENSDART00000075889 ENSDART00000148847 |
trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr14_+_46313135 | 0.32 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_+_65673885 | 0.32 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr10_+_2669405 | 0.31 |
ENSDART00000110202
|
ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr4_+_76705830 | 0.31 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr22_-_17631675 | 0.31 |
ENSDART00000132565
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr1_-_47161996 | 0.31 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr9_+_24088062 | 0.31 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr7_+_27603211 | 0.31 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr18_-_22713402 | 0.31 |
ENSDART00000145871
|
si:ch73-113g13.2
|
si:ch73-113g13.2 |
| chr1_-_10647484 | 0.31 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr19_-_657439 | 0.31 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr13_-_438705 | 0.31 |
ENSDART00000082142
|
CU570800.1
|
|
| chr15_-_17813680 | 0.30 |
ENSDART00000158556
|
CT573342.2
|
|
| chr16_+_35870456 | 0.30 |
ENSDART00000184321
|
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr6_-_54180699 | 0.30 |
ENSDART00000045901
|
rps10
|
ribosomal protein S10 |
| chr3_-_45848257 | 0.30 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr5_+_12515402 | 0.30 |
ENSDART00000103278
|
zgc:171242
|
zgc:171242 |
| chr4_+_15944245 | 0.30 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr16_+_7653905 | 0.30 |
ENSDART00000081431
|
popdc3
|
popeye domain containing 3 |
| chr16_+_35916371 | 0.30 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr1_+_40802454 | 0.30 |
ENSDART00000193568
|
cpz
|
carboxypeptidase Z |
| chr17_+_8799451 | 0.30 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr25_+_5044780 | 0.30 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr21_+_26071874 | 0.29 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr13_-_50341004 | 0.29 |
ENSDART00000038120
|
cacul1
|
CDK2 associated cullin domain 1 |
| chr1_-_10647307 | 0.29 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr9_-_51563575 | 0.29 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr10_+_10313024 | 0.29 |
ENSDART00000142895
ENSDART00000129952 |
urm1
|
ubiquitin related modifier 1 |
| chr11_-_40147032 | 0.29 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr1_-_513762 | 0.29 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr18_+_48793536 | 0.28 |
ENSDART00000168937
|
bmp16
|
bone morphogenetic protein 16 |
| chr15_+_14165474 | 0.28 |
ENSDART00000161891
ENSDART00000159757 |
clca5.2
|
chloride channel accessory 5, tandem duplicate 2 |
| chr16_+_29509133 | 0.28 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr14_+_923803 | 0.28 |
ENSDART00000111141
|
myoz3a
|
myozenin 3a |
| chr11_+_6136220 | 0.28 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr14_-_11456724 | 0.28 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr14_-_34059681 | 0.28 |
ENSDART00000003993
|
itk
|
IL2 inducible T cell kinase |
| chr19_+_31585341 | 0.28 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr8_-_45279411 | 0.28 |
ENSDART00000175207
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr6_+_43450221 | 0.28 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr10_+_40629616 | 0.28 |
ENSDART00000147476
|
CR396590.9
|
|
| chr2_+_48073972 | 0.27 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr6_+_53288018 | 0.27 |
ENSDART00000170319
|
cysltr3
|
cysteinyl leukotriene receptor 3 |
| chr1_-_12027 | 0.27 |
ENSDART00000164359
|
rpl24
|
ribosomal protein L24 |
| chr22_+_5532003 | 0.27 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr11_+_42585138 | 0.27 |
ENSDART00000019008
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr7_+_41295974 | 0.27 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr4_-_12477224 | 0.27 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr1_-_43684584 | 0.27 |
ENSDART00000074609
|
suclg1
|
succinate-CoA ligase, alpha subunit |
| chr1_+_40801696 | 0.27 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr5_+_1933131 | 0.27 |
ENSDART00000061693
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr3_-_32275975 | 0.27 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr16_+_17715243 | 0.27 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr23_-_19682971 | 0.27 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr1_-_6028876 | 0.26 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr17_+_29276544 | 0.26 |
ENSDART00000049399
|
ankrd9
|
ankyrin repeat domain 9 |
| chr12_+_13319132 | 0.26 |
ENSDART00000049167
|
psme2
|
proteasome activator subunit 2 |
| chr16_-_17072440 | 0.26 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr20_-_22778394 | 0.26 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr13_+_30692669 | 0.26 |
ENSDART00000187818
|
CR762483.1
|
|
| chr16_+_16977786 | 0.26 |
ENSDART00000043173
ENSDART00000132150 |
rpl18
|
ribosomal protein L18 |
| chr4_+_391297 | 0.26 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr16_+_19637384 | 0.26 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr7_+_32693890 | 0.26 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr16_+_36671064 | 0.26 |
ENSDART00000109703
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr6_-_54180516 | 0.26 |
ENSDART00000149945
|
rps10
|
ribosomal protein S10 |
| chr13_-_17723417 | 0.26 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr1_-_52461322 | 0.26 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr13_+_35637048 | 0.26 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr9_-_18911608 | 0.26 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr22_+_3153876 | 0.26 |
ENSDART00000163327
|
rpl36
|
ribosomal protein L36 |
| chr22_+_34694217 | 0.26 |
ENSDART00000111793
|
hykk.1
|
hydroxylysine kinase, tandem duplicate 1 |
| chr12_+_47663419 | 0.26 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr14_+_46313396 | 0.26 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr14_-_26536504 | 0.26 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr2_+_3502430 | 0.26 |
ENSDART00000113395
|
SERP1
|
zgc:92744 |
| chr13_+_40686133 | 0.25 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr17_-_5426458 | 0.25 |
ENSDART00000157937
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr17_-_7792376 | 0.25 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr9_-_15424639 | 0.25 |
ENSDART00000124346
|
fn1a
|
fibronectin 1a |
| chr15_-_5016039 | 0.25 |
ENSDART00000156458
|
defbl3
|
defensin, beta-like 3 |
| chr11_+_17984167 | 0.25 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr17_+_23730089 | 0.25 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr2_-_51757328 | 0.25 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr10_-_24759616 | 0.25 |
ENSDART00000079528
|
ilk
|
integrin-linked kinase |
| chr3_-_29870848 | 0.25 |
ENSDART00000186457
|
rpl3
|
ribosomal protein L3 |
| chr24_+_20927135 | 0.25 |
ENSDART00000144883
ENSDART00000131829 |
fam162a
|
family with sequence similarity 162, member A |
| chr23_-_1660708 | 0.25 |
ENSDART00000175138
|
CU693481.1
|
|
| chr11_+_17984354 | 0.25 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr24_-_6078222 | 0.25 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr7_-_8763123 | 0.25 |
ENSDART00000173189
ENSDART00000102567 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr2_+_52065884 | 0.25 |
ENSDART00000146418
|
shda
|
Src homology 2 domain containing transforming protein D, a |
| chr21_-_11575626 | 0.25 |
ENSDART00000133171
|
cast
|
calpastatin |
| chr24_-_32582880 | 0.25 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr23_-_39784368 | 0.24 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr3_+_21225750 | 0.24 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr12_+_32199651 | 0.24 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr18_-_44285539 | 0.24 |
ENSDART00000137222
|
pimr179
|
Pim proto-oncogene, serine/threonine kinase, related 179 |
| chr25_+_186583 | 0.24 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr3_-_45848043 | 0.24 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr12_+_27127139 | 0.24 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr19_+_41169996 | 0.24 |
ENSDART00000048438
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr10_+_29259882 | 0.24 |
ENSDART00000180606
|
sytl2a
|
synaptotagmin-like 2a |
| chr16_-_26537103 | 0.24 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr13_-_1130096 | 0.24 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr6_-_1591002 | 0.24 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr9_-_9980704 | 0.24 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr10_+_9410304 | 0.24 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr2_-_31686353 | 0.24 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr23_+_44157682 | 0.24 |
ENSDART00000164474
ENSDART00000149928 |
si:ch73-106g13.1
|
si:ch73-106g13.1 |
| chr5_-_54197084 | 0.24 |
ENSDART00000163640
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr7_-_6357952 | 0.23 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr16_+_31921812 | 0.23 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr22_+_15315655 | 0.23 |
ENSDART00000141249
|
sult3st3
|
sulfotransferase family 3, cytosolic sulfotransferase 3 |
| chr3_-_3428938 | 0.23 |
ENSDART00000179811
ENSDART00000115282 ENSDART00000192263 ENSDART00000046454 |
A2ML1 (1 of many)
|
zgc:171445 |
Network of associatons between targets according to the STRING database.
First level regulatory network of barx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.5 | GO:0061217 | regulation of neurotransmitter uptake(GO:0051580) regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.7 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.2 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.1 | 0.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.4 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.3 | GO:0000480 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.5 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.4 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.2 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.2 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.7 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0002902 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.2 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.3 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.2 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.1 | 0.2 | GO:0030237 | female sex determination(GO:0030237) |
| 0.1 | 0.2 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.2 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.4 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:2000648 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.2 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0006953 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0034650 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 1.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.4 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.4 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:2000379 | positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.0 | 0.2 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.8 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.0 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.0 | 0.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.7 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.4 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.0 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.3 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 0.1 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.0 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.2 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 3.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.9 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.0 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.4 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.5 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.2 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.2 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.2 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.2 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.2 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.2 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.3 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.2 | GO:0015440 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 1.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 3.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |