Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
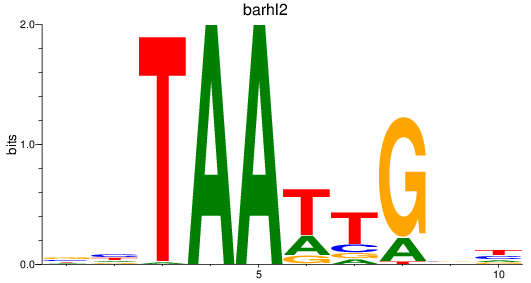
Results for barhl2
Z-value: 0.77
Transcription factors associated with barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barhl2
|
ENSDARG00000104361 | BarH-like homeobox 2 |
|
barhl2
|
ENSDARG00000114681 | BarH-like homeobox 2 |
|
barhl2
|
ENSDARG00000115723 | BarH-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barhl2 | dr11_v1_chr6_+_24817852_24817852 | -0.88 | 4.8e-02 | Click! |
Activity profile of barhl2 motif
Sorted Z-values of barhl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_52437056 | 0.54 |
ENSDART00000138337
|
si:ch211-217k17.12
|
si:ch211-217k17.12 |
| chr12_+_20352400 | 0.43 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr24_-_4765740 | 0.40 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr15_+_34699585 | 0.40 |
ENSDART00000017569
|
tnfb
|
tumor necrosis factor b (TNF superfamily, member 2) |
| chr4_+_73606482 | 0.36 |
ENSDART00000150765
|
si:ch211-165i18.2
|
si:ch211-165i18.2 |
| chr21_+_25777425 | 0.35 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr15_-_2640966 | 0.35 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr1_+_40802454 | 0.34 |
ENSDART00000193568
|
cpz
|
carboxypeptidase Z |
| chr1_-_19845378 | 0.34 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr7_+_13988075 | 0.34 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr16_-_26140768 | 0.33 |
ENSDART00000143960
|
cd79a
|
CD79a molecule, immunoglobulin-associated alpha |
| chr14_+_14568437 | 0.33 |
ENSDART00000164749
|
pcdh20
|
protocadherin 20 |
| chr8_+_23738122 | 0.33 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr11_+_6136220 | 0.32 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr1_-_513762 | 0.32 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr3_-_20091964 | 0.31 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr6_-_8362419 | 0.31 |
ENSDART00000142752
ENSDART00000135810 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr19_+_10855158 | 0.31 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr23_+_38171186 | 0.30 |
ENSDART00000148188
|
zgc:112994
|
zgc:112994 |
| chr5_-_34993242 | 0.30 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr17_+_25849332 | 0.30 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr22_+_10066846 | 0.29 |
ENSDART00000105916
ENSDART00000105914 ENSDART00000132480 |
si:ch211-222k6.3
|
si:ch211-222k6.3 |
| chr5_-_42272517 | 0.29 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr8_-_38201415 | 0.29 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr10_-_13343831 | 0.28 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr1_-_33645967 | 0.28 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr20_+_38257766 | 0.28 |
ENSDART00000147485
ENSDART00000149160 |
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr16_+_7697878 | 0.28 |
ENSDART00000104176
ENSDART00000172977 |
ccr11.1
|
chemokine (C-C motif) receptor 11.1 |
| chr20_-_54924593 | 0.28 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr16_-_23797570 | 0.28 |
ENSDART00000077834
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr11_-_22303678 | 0.28 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr23_-_19715557 | 0.28 |
ENSDART00000143764
|
rpl10
|
ribosomal protein L10 |
| chr9_+_14010823 | 0.27 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr5_+_9246458 | 0.27 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr3_-_32541033 | 0.27 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr16_+_23398369 | 0.27 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr11_-_7261717 | 0.27 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr22_-_24297510 | 0.27 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr5_-_38820046 | 0.27 |
ENSDART00000182886
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr8_+_13389115 | 0.26 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr7_+_4474880 | 0.26 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr18_+_3037998 | 0.25 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr5_+_29794058 | 0.25 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr23_+_36095260 | 0.25 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr12_-_28983584 | 0.25 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
| chr5_+_6670945 | 0.25 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr6_+_28877306 | 0.25 |
ENSDART00000065137
ENSDART00000123189 ENSDART00000065135 ENSDART00000181512 ENSDART00000130799 |
tp63
|
tumor protein p63 |
| chr11_+_11303458 | 0.24 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr10_-_22127942 | 0.24 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr3_-_32071468 | 0.24 |
ENSDART00000121979
|
BX511080.1
|
|
| chr16_-_9694822 | 0.24 |
ENSDART00000168748
|
COLEC10
|
si:ch211-93i7.4 |
| chr11_-_30634286 | 0.24 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr10_+_40629616 | 0.24 |
ENSDART00000147476
|
CR396590.9
|
|
| chr13_+_829585 | 0.23 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr11_+_18130300 | 0.23 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr13_-_50139916 | 0.23 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr3_-_29928695 | 0.23 |
ENSDART00000151275
ENSDART00000151083 |
grna
|
granulin a |
| chr5_+_27897504 | 0.23 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr13_-_11984867 | 0.23 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr12_-_46112892 | 0.23 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr11_+_18157260 | 0.23 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr2_+_50999477 | 0.23 |
ENSDART00000190111
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr5_-_30615901 | 0.22 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr6_+_41191482 | 0.22 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr17_-_20118145 | 0.22 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr6_-_7720332 | 0.22 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr19_+_43297546 | 0.22 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr16_-_7443388 | 0.22 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr10_-_44560165 | 0.22 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr22_+_3156386 | 0.22 |
ENSDART00000161212
|
rpl36
|
ribosomal protein L36 |
| chr21_-_11654422 | 0.21 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr7_+_17443567 | 0.21 |
ENSDART00000060383
|
nitr2b
|
novel immune-type receptor 2b |
| chr23_+_19813677 | 0.21 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr20_-_23426339 | 0.21 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_-_10006158 | 0.21 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr11_-_40457325 | 0.21 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr23_+_32029304 | 0.21 |
ENSDART00000185217
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr17_-_30635298 | 0.21 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr10_+_29840725 | 0.21 |
ENSDART00000127268
|
BX120005.1
|
|
| chr15_-_951118 | 0.21 |
ENSDART00000171427
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr22_-_10541372 | 0.20 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr11_+_13166750 | 0.20 |
ENSDART00000169961
|
mob3c
|
MOB kinase activator 3C |
| chr22_-_14272699 | 0.20 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr1_-_56032619 | 0.20 |
ENSDART00000143793
|
c3a.4
|
complement component c3a, duplicate 4 |
| chr16_-_26731928 | 0.20 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr7_+_17816006 | 0.20 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr10_-_7785930 | 0.20 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr20_+_26880668 | 0.20 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr22_-_20695237 | 0.20 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr9_-_6991650 | 0.19 |
ENSDART00000081718
|
slc9a2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr5_-_69620722 | 0.19 |
ENSDART00000097248
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr23_-_19230627 | 0.19 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr4_-_17669881 | 0.19 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr15_+_22394074 | 0.19 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr1_-_18811517 | 0.19 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr18_+_9171778 | 0.19 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr14_-_32884138 | 0.19 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr21_-_17296789 | 0.19 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr21_+_11560153 | 0.19 |
ENSDART00000065842
|
cd8a
|
CD8a molecule |
| chr17_-_23616626 | 0.19 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr17_+_16046314 | 0.19 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_+_31039091 | 0.19 |
ENSDART00000060005
|
rpl32
|
ribosomal protein L32 |
| chr23_+_9220436 | 0.18 |
ENSDART00000033663
ENSDART00000139870 |
rps21
|
ribosomal protein S21 |
| chr5_-_72390259 | 0.18 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr9_+_23714406 | 0.18 |
ENSDART00000189445
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr11_-_40519886 | 0.18 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr24_-_25166720 | 0.18 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr17_+_43867889 | 0.18 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr25_-_16724913 | 0.18 |
ENSDART00000157075
|
galnt8a.2
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 2 |
| chr16_+_41826584 | 0.18 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr20_+_2039518 | 0.18 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr16_+_42471455 | 0.18 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr22_+_19538626 | 0.18 |
ENSDART00000189174
|
BX936298.2
|
|
| chr7_-_20158380 | 0.18 |
ENSDART00000142891
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr9_-_21912227 | 0.18 |
ENSDART00000145576
|
lmo7a
|
LIM domain 7a |
| chr9_+_1313418 | 0.18 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr18_-_25568994 | 0.18 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr16_+_26732086 | 0.18 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr16_-_48430272 | 0.18 |
ENSDART00000005927
|
rad21a
|
RAD21 cohesin complex component a |
| chr2_-_55779927 | 0.18 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr14_+_26247319 | 0.17 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr6_+_39098397 | 0.17 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr6_+_19948043 | 0.17 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr13_+_835390 | 0.17 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr12_-_11349899 | 0.17 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr18_+_29898955 | 0.17 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr25_+_15939275 | 0.17 |
ENSDART00000126641
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr2_-_45510699 | 0.17 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr5_+_9246018 | 0.17 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr2_-_49985351 | 0.17 |
ENSDART00000145888
|
ccr12a
|
chemokine (C-C motif) receptor 12a |
| chr18_-_18942098 | 0.17 |
ENSDART00000100458
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr24_-_34680956 | 0.17 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr11_-_44801968 | 0.17 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr9_-_24031461 | 0.17 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr8_-_7847921 | 0.17 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr21_+_382400 | 0.17 |
ENSDART00000190785
ENSDART00000124615 |
rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr19_+_5640504 | 0.16 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr8_-_24252933 | 0.16 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr21_-_19919020 | 0.16 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr3_+_26244353 | 0.16 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr20_-_33675676 | 0.16 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr7_-_64971839 | 0.16 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr4_-_77218637 | 0.16 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr22_+_1092479 | 0.16 |
ENSDART00000170119
|
guca1e
|
guanylate cyclase activator 1e |
| chr18_-_34549721 | 0.16 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr5_+_61998641 | 0.16 |
ENSDART00000137110
|
selenow1
|
selenoprotein W, 1 |
| chr19_-_20403507 | 0.16 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr8_+_45334255 | 0.16 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr11_+_25539698 | 0.16 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr8_+_41037541 | 0.15 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr11_+_24716837 | 0.15 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr7_-_20464468 | 0.15 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr4_+_9508505 | 0.15 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr8_-_46486009 | 0.15 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr25_-_18435481 | 0.15 |
ENSDART00000004771
|
poc1b
|
POC1 centriolar protein B |
| chr7_+_38510197 | 0.15 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr21_+_25236297 | 0.15 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr19_+_11217279 | 0.15 |
ENSDART00000181859
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr19_-_20403845 | 0.15 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr3_+_5490104 | 0.15 |
ENSDART00000170191
|
FQ311879.1
|
|
| chr9_+_21165017 | 0.15 |
ENSDART00000145933
ENSDART00000142985 |
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr13_-_40401870 | 0.15 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr18_+_27300700 | 0.15 |
ENSDART00000140444
|
plekha7a
|
pleckstrin homology domain containing, family A member 7a |
| chr16_+_50972803 | 0.15 |
ENSDART00000178189
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr7_-_7972445 | 0.15 |
ENSDART00000182687
ENSDART00000191946 |
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr22_+_31025096 | 0.15 |
ENSDART00000185953
|
zmp:0000000735
|
zmp:0000000735 |
| chr7_+_2236317 | 0.15 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr22_+_30543437 | 0.15 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr1_+_513986 | 0.15 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr1_+_40801696 | 0.15 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr2_+_3201345 | 0.14 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr9_+_30475563 | 0.14 |
ENSDART00000133118
|
gja5a
|
gap junction protein, alpha 5a |
| chr17_-_31695217 | 0.14 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr24_+_36414829 | 0.14 |
ENSDART00000062699
|
si:ch211-253b1.3
|
si:ch211-253b1.3 |
| chr3_+_22381955 | 0.14 |
ENSDART00000185315
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr14_+_23709134 | 0.14 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr8_+_22472584 | 0.14 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr9_-_35633827 | 0.14 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr5_+_29652513 | 0.14 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr3_+_31093455 | 0.14 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr18_-_26894732 | 0.14 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr24_+_12835935 | 0.14 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr24_-_28245872 | 0.14 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr4_+_28997595 | 0.14 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr6_-_10927766 | 0.13 |
ENSDART00000134327
|
ccr7
|
chemokine (C-C motif) receptor 7 |
| chr3_+_27798094 | 0.13 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr21_+_26657404 | 0.13 |
ENSDART00000129035
ENSDART00000186550 |
prdx5
|
peroxiredoxin 5 |
| chr17_+_22530820 | 0.13 |
ENSDART00000089916
|
edaradd
|
EDAR-associated death domain |
| chr19_+_7045033 | 0.13 |
ENSDART00000146579
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr18_+_19120984 | 0.13 |
ENSDART00000141501
|
si:dkey-242h9.3
|
si:dkey-242h9.3 |
| chr2_+_16780643 | 0.13 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr3_-_42125655 | 0.13 |
ENSDART00000040753
|
nudt1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr19_+_31904836 | 0.13 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr9_-_10778615 | 0.13 |
ENSDART00000182577
|
CABZ01053619.1
|
|
| chr17_+_16046132 | 0.13 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_+_39663987 | 0.13 |
ENSDART00000184614
ENSDART00000184573 ENSDART00000183127 |
epn2
|
epsin 2 |
| chr6_+_42475730 | 0.13 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr15_+_24795473 | 0.13 |
ENSDART00000139689
ENSDART00000141033 ENSDART00000100746 ENSDART00000135677 |
gosr1
|
golgi SNAP receptor complex member 1 |
| chr10_-_17988779 | 0.13 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr9_-_53666031 | 0.13 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of barhl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.3 | GO:0051000 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.1 | 0.2 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.2 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.3 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.2 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) positive regulation of Golgi to plasma membrane protein transport(GO:0042998) regulation of establishment of protein localization to plasma membrane(GO:0090003) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0045226 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.2 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.1 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.3 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.8 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.0 | 0.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.0 | 0.1 | GO:0072679 | chemokine production(GO:0032602) negative T cell selection(GO:0043383) thymocyte migration(GO:0072679) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.2 | GO:0006971 | hypotonic response(GO:0006971) hypotonic salinity response(GO:0042539) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) lateral line ganglion development(GO:0048890) |
| 0.0 | 0.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.2 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.4 | GO:0030250 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.0 | 0.1 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0015217 | ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |