Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
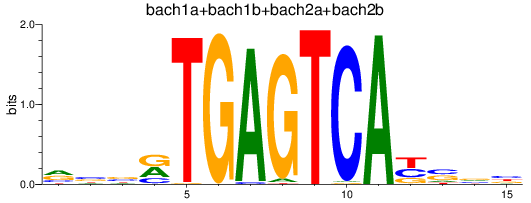
Results for bach1a+bach1b+bach2a+bach2b
Z-value: 1.34
Transcription factors associated with bach1a+bach1b+bach2a+bach2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bach1b
|
ENSDARG00000002196 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
|
bach2b
|
ENSDARG00000004074 | BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
|
bach2a
|
ENSDARG00000036569 | BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
|
bach1a
|
ENSDARG00000062553 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bach2a | dr11_v1_chr17_+_15674052_15674052 | 0.95 | 1.4e-02 | Click! |
| bach2b | dr11_v1_chr20_-_24022558_24022558 | 0.84 | 7.6e-02 | Click! |
| bach1a | dr11_v1_chr15_-_8191992_8191992 | 0.76 | 1.4e-01 | Click! |
| bach1b | dr11_v1_chr10_+_25369254_25369254 | -0.58 | 3.0e-01 | Click! |
Activity profile of bach1a+bach1b+bach2a+bach2b motif
Sorted Z-values of bach1a+bach1b+bach2a+bach2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_21832441 | 2.37 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr19_-_103289 | 2.00 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr1_+_31113951 | 1.85 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr14_+_34486629 | 1.75 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr2_+_47582681 | 1.66 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr21_-_30545121 | 1.48 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr2_+_47582488 | 1.47 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr2_+_47581997 | 1.40 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr16_+_14033121 | 1.37 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr1_-_53750522 | 1.35 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr20_+_34915945 | 1.35 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr12_-_10315039 | 1.32 |
ENSDART00000152680
|
pyyb
|
peptide YYb |
| chr3_+_7617353 | 1.29 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr1_+_12766351 | 1.21 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr1_-_40227166 | 1.20 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr21_+_3093419 | 1.20 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr2_-_155270 | 1.19 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr7_+_23875269 | 1.15 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr8_+_7359294 | 1.12 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr23_-_15878879 | 1.11 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr1_+_41690402 | 1.10 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr16_-_12984631 | 1.07 |
ENSDART00000184863
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr15_+_28989573 | 1.06 |
ENSDART00000076648
|
clip3
|
CAP-GLY domain containing linker protein 3 |
| chr5_-_22501663 | 1.05 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr1_+_46598764 | 1.04 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr19_-_5805923 | 1.02 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr2_+_24177006 | 1.01 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr3_+_45401472 | 0.98 |
ENSDART00000156693
|
baiap3
|
BAI1-associated protein 3 |
| chr22_-_3595439 | 0.96 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr2_+_50862527 | 0.94 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr19_-_36675023 | 0.93 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr7_+_26649319 | 0.92 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr5_-_14564878 | 0.91 |
ENSDART00000160511
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr7_-_52963493 | 0.88 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr2_+_48288461 | 0.88 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr8_+_24861264 | 0.87 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr8_+_24854600 | 0.87 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr13_-_21701323 | 0.86 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr21_-_42100471 | 0.80 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr5_-_20678300 | 0.79 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr25_+_21324588 | 0.79 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr1_+_46598502 | 0.77 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr2_-_9544161 | 0.77 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr10_+_4235998 | 0.76 |
ENSDART00000169328
|
plppr1
|
phospholipid phosphatase related 1 |
| chr5_+_20147830 | 0.76 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr13_-_37545487 | 0.75 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr17_-_7861219 | 0.75 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr8_-_13211527 | 0.74 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr2_+_24177190 | 0.74 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr16_-_32837806 | 0.73 |
ENSDART00000003997
|
MCHR2
|
si:dkey-165n16.5 |
| chr2_+_20430366 | 0.72 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr1_+_41849152 | 0.72 |
ENSDART00000053685
|
smox
|
spermine oxidase |
| chr25_-_7999756 | 0.72 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr17_-_45247332 | 0.71 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr15_+_40188076 | 0.70 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr21_-_45613564 | 0.70 |
ENSDART00000160324
|
LO018363.1
|
|
| chr4_-_4535189 | 0.69 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr15_+_32867420 | 0.69 |
ENSDART00000159442
|
dclk1b
|
doublecortin-like kinase 1b |
| chr18_-_18850302 | 0.67 |
ENSDART00000131965
ENSDART00000167624 |
tgm2l
|
transglutaminase 2, like |
| chr19_+_24394560 | 0.67 |
ENSDART00000142506
|
si:dkey-81h8.1
|
si:dkey-81h8.1 |
| chr5_+_15350954 | 0.66 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr5_+_20035284 | 0.64 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr15_-_13254480 | 0.63 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr10_+_37145007 | 0.62 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr7_-_5487593 | 0.62 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr1_+_2190714 | 0.61 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr6_+_10534752 | 0.61 |
ENSDART00000090874
|
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr23_-_28239750 | 0.60 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr24_-_40009446 | 0.60 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr22_-_11954861 | 0.59 |
ENSDART00000131611
|
si:dkeyp-4h4.1
|
si:dkeyp-4h4.1 |
| chr14_-_34044369 | 0.59 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_9046805 | 0.59 |
ENSDART00000134224
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr24_-_21973365 | 0.58 |
ENSDART00000081204
ENSDART00000030592 |
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr16_+_34479064 | 0.57 |
ENSDART00000159965
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr19_+_10396042 | 0.57 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr2_+_42112503 | 0.57 |
ENSDART00000136022
|
crha
|
corticotropin releasing hormone a |
| chr12_-_17147473 | 0.55 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr18_+_3169579 | 0.55 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr3_+_20156956 | 0.55 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr5_+_59534286 | 0.54 |
ENSDART00000189341
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr18_+_9635178 | 0.53 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr15_-_30832171 | 0.52 |
ENSDART00000152414
ENSDART00000152790 |
msi2b
|
musashi RNA-binding protein 2b |
| chr4_-_1801519 | 0.52 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr5_+_44804791 | 0.51 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr17_-_45247151 | 0.51 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr10_-_32877348 | 0.51 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr22_+_24220937 | 0.49 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr19_-_10395683 | 0.49 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr11_+_19370717 | 0.49 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr21_-_30254185 | 0.49 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr18_+_3243292 | 0.48 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr24_+_26039464 | 0.48 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr3_-_51142942 | 0.47 |
ENSDART00000160789
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr12_-_35386910 | 0.47 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr19_-_7495006 | 0.46 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr10_-_27566481 | 0.45 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr2_+_9822319 | 0.45 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr11_-_3334248 | 0.44 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr25_-_11378623 | 0.44 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr25_+_18583877 | 0.44 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr13_+_43247936 | 0.43 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr11_+_19370447 | 0.43 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr24_-_5691956 | 0.42 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr23_+_45339684 | 0.42 |
ENSDART00000149410
ENSDART00000102441 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr17_+_19626479 | 0.41 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr14_-_6936731 | 0.40 |
ENSDART00000162196
|
clk4a
|
CDC-like kinase 4a |
| chr2_+_9821757 | 0.40 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr19_-_40776608 | 0.40 |
ENSDART00000179941
|
calcr
|
calcitonin receptor |
| chr14_-_493029 | 0.39 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr3_+_24986145 | 0.39 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr9_+_29323163 | 0.39 |
ENSDART00000184908
|
oxgr1b
|
oxoglutarate (alpha-ketoglutarate) receptor 1b |
| chr8_+_48966165 | 0.38 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr19_+_33093395 | 0.38 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr25_-_173165 | 0.38 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr19_+_23932259 | 0.38 |
ENSDART00000139040
|
si:dkey-222b8.1
|
si:dkey-222b8.1 |
| chr20_-_16849306 | 0.37 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr2_-_39759059 | 0.37 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr19_+_33093577 | 0.37 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr21_+_26620867 | 0.37 |
ENSDART00000176008
|
esrra
|
estrogen-related receptor alpha |
| chr20_-_16548912 | 0.36 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr14_-_44841335 | 0.36 |
ENSDART00000173011
|
GRXCR1
|
si:dkey-109l4.6 |
| chr11_+_11267829 | 0.36 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr16_+_13818500 | 0.36 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr20_-_21994901 | 0.36 |
ENSDART00000004984
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr4_+_1600034 | 0.35 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr18_+_15841449 | 0.35 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr5_+_44805028 | 0.34 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr14_-_17588345 | 0.34 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr6_+_43903209 | 0.34 |
ENSDART00000006435
|
gpr27
|
G protein-coupled receptor 27 |
| chr20_-_32446406 | 0.33 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr14_-_44841503 | 0.33 |
ENSDART00000179114
|
GRXCR1
|
si:dkey-109l4.6 |
| chr5_+_21931124 | 0.33 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr22_+_17399124 | 0.32 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr17_-_31044803 | 0.32 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr25_+_14697247 | 0.32 |
ENSDART00000180747
|
mpped2
|
metallophosphoesterase domain containing 2b |
| chr3_+_35498119 | 0.32 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr18_-_18849916 | 0.32 |
ENSDART00000142422
|
tgm2l
|
transglutaminase 2, like |
| chr16_+_13818743 | 0.32 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr18_-_15155962 | 0.31 |
ENSDART00000111333
|
mterf2
|
mitochondrial transcription termination factor 2 |
| chr5_-_42180205 | 0.31 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr4_-_77506362 | 0.30 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr15_-_11955485 | 0.30 |
ENSDART00000160286
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr9_+_7030016 | 0.30 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr2_-_7605121 | 0.29 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr11_+_11267493 | 0.28 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr18_-_13360106 | 0.28 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr4_+_45274792 | 0.28 |
ENSDART00000150295
|
znf1138
|
zinc finger protein 1138 |
| chr16_-_21181128 | 0.28 |
ENSDART00000133403
|
pde11al
|
phosphodiesterase 11a, like |
| chr10_+_31222433 | 0.28 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr19_+_4139065 | 0.27 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr19_-_31035155 | 0.27 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr5_-_19052184 | 0.25 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr2_-_3038904 | 0.25 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr2_-_37837056 | 0.25 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr7_-_19638319 | 0.24 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr8_-_34402839 | 0.24 |
ENSDART00000180173
ENSDART00000191588 ENSDART00000186376 |
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr1_-_10577945 | 0.24 |
ENSDART00000179237
ENSDART00000040502 ENSDART00000186876 |
trpc5a
|
transient receptor potential cation channel, subfamily C, member 5a |
| chr1_+_58677434 | 0.24 |
ENSDART00000158488
|
LO017784.1
|
|
| chr3_+_24197934 | 0.24 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr11_+_34760628 | 0.24 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr4_-_73787702 | 0.23 |
ENSDART00000136328
ENSDART00000150546 |
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr11_-_22371105 | 0.23 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr19_+_551963 | 0.23 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr7_+_22688781 | 0.23 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr5_-_42123794 | 0.23 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr15_+_24704798 | 0.22 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr7_-_29723761 | 0.22 |
ENSDART00000173560
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr24_+_80653 | 0.21 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr1_+_56447107 | 0.21 |
ENSDART00000091924
|
CABZ01059392.1
|
|
| chr8_+_43053519 | 0.21 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr13_+_1430094 | 0.21 |
ENSDART00000169888
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr10_+_38775959 | 0.21 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr12_+_47280839 | 0.21 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr5_+_34549845 | 0.21 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr4_+_68087932 | 0.21 |
ENSDART00000150494
|
znf1096
|
zinc finger protein 1096 |
| chr22_-_6562618 | 0.20 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr18_+_17537344 | 0.20 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr18_-_8396192 | 0.20 |
ENSDART00000193830
|
BEND7
|
si:ch211-220f12.4 |
| chr3_-_24205339 | 0.20 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
| chr10_+_2529037 | 0.19 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr10_+_38775408 | 0.19 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr9_+_28140089 | 0.19 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr17_+_58211 | 0.19 |
ENSDART00000157642
|
si:ch1073-209e23.1
|
si:ch1073-209e23.1 |
| chr1_+_52137528 | 0.19 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr18_+_15182965 | 0.19 |
ENSDART00000137018
|
si:ch73-62b13.1
|
si:ch73-62b13.1 |
| chr4_+_40063572 | 0.19 |
ENSDART00000146710
|
BX649521.1
|
|
| chr16_+_48714048 | 0.19 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr22_+_36547670 | 0.18 |
ENSDART00000125991
ENSDART00000186372 |
armc9
|
armadillo repeat containing 9 |
| chr17_+_7595356 | 0.18 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr4_+_76431531 | 0.18 |
ENSDART00000174278
|
FP074874.1
|
|
| chr5_-_37157510 | 0.18 |
ENSDART00000166710
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr9_-_23181062 | 0.18 |
ENSDART00000192578
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr25_+_336503 | 0.17 |
ENSDART00000160395
|
CU929262.1
|
|
| chr12_-_3077395 | 0.17 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_36746837 | 0.17 |
ENSDART00000189276
ENSDART00000085228 |
pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr6_-_35309661 | 0.17 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr1_-_22726233 | 0.17 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr9_+_41024973 | 0.17 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr7_-_5396154 | 0.17 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr17_+_28675120 | 0.16 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bach1a+bach1b+bach2a+bach2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0015824 | proline transport(GO:0015824) |
| 0.5 | 2.0 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 1.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 4.7 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.9 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.5 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.7 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.3 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 2.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.2 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 1.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.1 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 1.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 0.3 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 1.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 3.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.9 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.4 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.5 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 1.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 3.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 2.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 4.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 2.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.9 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.7 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.5 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.5 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.7 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 3.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |