Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
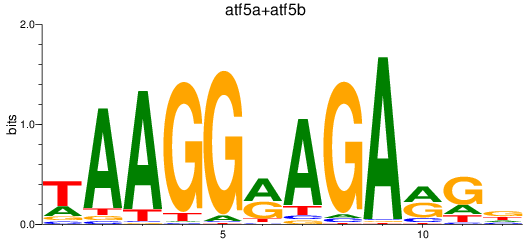
Results for atf5a+atf5b
Z-value: 1.67
Transcription factors associated with atf5a+atf5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf5a
|
ENSDARG00000068096 | activating transcription factor 5a |
|
atf5b
|
ENSDARG00000077785 | activating transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf5b | dr11_v1_chr15_-_17869115_17869115 | 0.85 | 7.0e-02 | Click! |
| atf5a | dr11_v1_chr5_-_30418636_30418636 | 0.54 | 3.5e-01 | Click! |
Activity profile of atf5a+atf5b motif
Sorted Z-values of atf5a+atf5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_26521970 | 1.88 |
ENSDART00000143712
|
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr19_-_21832441 | 1.67 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr9_+_38962017 | 1.53 |
ENSDART00000140436
|
map2
|
microtubule-associated protein 2 |
| chr21_-_22115136 | 1.37 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr3_-_16227490 | 1.37 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr9_+_31282161 | 1.24 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr16_+_29303971 | 1.22 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr9_-_27648683 | 1.21 |
ENSDART00000017292
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr7_-_28147838 | 1.19 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr3_-_16227683 | 1.16 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr18_+_17786548 | 1.14 |
ENSDART00000189493
ENSDART00000146133 |
ZNF423
|
si:ch211-216l23.1 |
| chr21_+_13908858 | 1.13 |
ENSDART00000148199
|
stxbp1a
|
syntaxin binding protein 1a |
| chr24_+_34606966 | 1.10 |
ENSDART00000105477
|
lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr7_+_26138240 | 1.09 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr18_+_22793465 | 1.08 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr1_-_5746030 | 1.07 |
ENSDART00000150863
|
nrp2a
|
neuropilin 2a |
| chr3_+_29714775 | 1.06 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr19_+_30662529 | 1.06 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr5_+_25952340 | 1.05 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr13_-_35051897 | 1.03 |
ENSDART00000129559
|
btbd3b
|
BTB (POZ) domain containing 3b |
| chr9_-_35069645 | 1.00 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr17_-_20711735 | 0.98 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr6_-_52400896 | 0.96 |
ENSDART00000187624
|
mmp24
|
matrix metallopeptidase 24 |
| chr3_-_30123113 | 0.94 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr16_-_24518027 | 0.93 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr17_-_37214196 | 0.91 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr24_-_1341543 | 0.91 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr13_-_33007781 | 0.90 |
ENSDART00000183671
ENSDART00000179859 |
rbm25a
|
RNA binding motif protein 25a |
| chr21_+_13353263 | 0.86 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr13_-_21739142 | 0.86 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr18_+_7283283 | 0.85 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr3_-_46818001 | 0.83 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_+_22793743 | 0.83 |
ENSDART00000150106
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr3_-_46817838 | 0.83 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_-_23875370 | 0.77 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_+_14225048 | 0.77 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr18_-_38088099 | 0.77 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr10_-_43568239 | 0.77 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr7_+_25323742 | 0.76 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr5_-_64213253 | 0.76 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr13_-_23007813 | 0.74 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr18_-_38087875 | 0.74 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr15_+_22267847 | 0.73 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr14_+_14224730 | 0.73 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr19_-_3931917 | 0.72 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr2_-_34555945 | 0.69 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr20_-_23026223 | 0.68 |
ENSDART00000015755
|
rasl11b
|
RAS-like, family 11, member B |
| chr12_+_8373525 | 0.68 |
ENSDART00000152180
|
arid5b
|
AT-rich interaction domain 5B |
| chr18_-_29977431 | 0.68 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr16_-_9802998 | 0.66 |
ENSDART00000154217
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr3_-_26341959 | 0.66 |
ENSDART00000169344
ENSDART00000142878 ENSDART00000087196 |
zgc:153240
|
zgc:153240 |
| chr23_-_35069805 | 0.64 |
ENSDART00000087219
|
BX294434.1
|
|
| chr3_+_15358459 | 0.62 |
ENSDART00000141808
|
sh2b1
|
SH2B adaptor protein 1 |
| chr5_+_58492699 | 0.62 |
ENSDART00000181584
|
FQ378016.1
|
|
| chr5_-_33215261 | 0.61 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr1_+_32521469 | 0.60 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr3_+_22905341 | 0.60 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr17_-_37054959 | 0.59 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr24_+_9372292 | 0.59 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr16_-_44649053 | 0.59 |
ENSDART00000184807
|
CR925804.2
|
|
| chr16_+_10346277 | 0.57 |
ENSDART00000081092
|
si:dkeyp-77h1.4
|
si:dkeyp-77h1.4 |
| chr22_-_30935510 | 0.57 |
ENSDART00000133335
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr21_-_17482465 | 0.56 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr9_+_25775816 | 0.55 |
ENSDART00000127834
ENSDART00000189994 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr5_-_48307804 | 0.55 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr16_-_13004166 | 0.54 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr5_-_67911111 | 0.53 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr3_+_15358625 | 0.53 |
ENSDART00000143280
|
sh2b1
|
SH2B adaptor protein 1 |
| chr7_+_20019125 | 0.53 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr13_+_28854438 | 0.53 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr17_-_20849879 | 0.52 |
ENSDART00000088100
ENSDART00000149630 |
ank3b
|
ankyrin 3b |
| chr18_+_28988373 | 0.52 |
ENSDART00000018685
|
syt9a
|
synaptotagmin IXa |
| chr18_-_23875219 | 0.50 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_-_28679135 | 0.49 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr17_-_37052622 | 0.49 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr14_-_44841335 | 0.49 |
ENSDART00000173011
|
GRXCR1
|
si:dkey-109l4.6 |
| chr11_-_22916641 | 0.48 |
ENSDART00000080201
ENSDART00000154813 |
mdm4
|
MDM4, p53 regulator |
| chr3_-_37476278 | 0.47 |
ENSDART00000083442
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr15_+_36977208 | 0.47 |
ENSDART00000183625
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr5_+_42467867 | 0.47 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr23_-_27607039 | 0.46 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr5_+_22791686 | 0.46 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr11_-_44030962 | 0.46 |
ENSDART00000171910
|
FP016005.1
|
|
| chr16_-_29146624 | 0.46 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr16_+_39159752 | 0.46 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr20_+_23676649 | 0.44 |
ENSDART00000035239
|
nek1
|
NIMA-related kinase 1 |
| chr14_-_44841503 | 0.43 |
ENSDART00000179114
|
GRXCR1
|
si:dkey-109l4.6 |
| chr24_-_11309477 | 0.42 |
ENSDART00000137257
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr20_-_26421112 | 0.42 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr16_-_34338537 | 0.42 |
ENSDART00000142223
|
hivep3b
|
human immunodeficiency virus type I enhancer binding protein 3b |
| chr23_-_27692717 | 0.41 |
ENSDART00000053878
ENSDART00000145028 |
IKZF4
|
si:dkey-166n8.9 |
| chr18_+_22174630 | 0.41 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr18_-_50152689 | 0.40 |
ENSDART00000006078
|
loxl1
|
lysyl oxidase-like 1 |
| chr16_+_25259313 | 0.40 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr20_-_26420939 | 0.39 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr17_+_42027969 | 0.39 |
ENSDART00000147563
|
kiz
|
kizuna centrosomal protein |
| chr5_-_40178092 | 0.38 |
ENSDART00000146664
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr4_-_8902406 | 0.38 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr17_+_25432449 | 0.38 |
ENSDART00000150195
|
srrm1
|
serine/arginine repetitive matrix 1 |
| chr18_+_34362608 | 0.37 |
ENSDART00000131478
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr12_+_27024676 | 0.36 |
ENSDART00000153104
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr21_+_33249478 | 0.36 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr1_+_16573982 | 0.35 |
ENSDART00000166317
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr1_+_52130213 | 0.35 |
ENSDART00000018817
|
rnf11a
|
ring finger protein 11a |
| chr6_+_36807861 | 0.34 |
ENSDART00000161708
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr22_+_3045495 | 0.34 |
ENSDART00000164061
|
LO017843.1
|
|
| chr16_-_24135508 | 0.34 |
ENSDART00000171819
ENSDART00000103176 |
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr14_+_8127893 | 0.33 |
ENSDART00000169091
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr6_-_34008827 | 0.33 |
ENSDART00000191183
ENSDART00000003701 ENSDART00000192502 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr19_-_10214264 | 0.33 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr9_-_41401564 | 0.33 |
ENSDART00000059628
|
nab1b
|
NGFI-A binding protein 1b (EGR1 binding protein 1) |
| chr13_+_23214100 | 0.32 |
ENSDART00000163393
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr5_+_45140914 | 0.32 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_41498697 | 0.31 |
ENSDART00000114230
|
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr17_+_38262408 | 0.30 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr21_-_26691959 | 0.30 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_-_29902187 | 0.29 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr18_+_17786710 | 0.29 |
ENSDART00000190203
ENSDART00000187095 ENSDART00000083296 |
ZNF423
|
si:ch211-216l23.1 |
| chr21_-_38031038 | 0.29 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr4_+_5180650 | 0.28 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr1_-_5745420 | 0.28 |
ENSDART00000166779
|
nrp2a
|
neuropilin 2a |
| chr12_-_3778848 | 0.28 |
ENSDART00000152128
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr10_+_29199172 | 0.28 |
ENSDART00000148828
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr7_-_20758825 | 0.27 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr1_+_41498188 | 0.27 |
ENSDART00000191934
ENSDART00000146310 |
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr14_-_24110251 | 0.26 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr5_-_19052184 | 0.24 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr9_-_21230999 | 0.23 |
ENSDART00000150160
ENSDART00000102147 |
pla1a
|
phospholipase A1 member A |
| chr2_+_30916188 | 0.23 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr12_-_31995840 | 0.23 |
ENSDART00000112881
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr22_+_30089054 | 0.23 |
ENSDART00000185369
|
add3a
|
adducin 3 (gamma) a |
| chr14_-_24110062 | 0.22 |
ENSDART00000177062
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr25_-_16832705 | 0.22 |
ENSDART00000171336
ENSDART00000189396 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr11_+_26476153 | 0.22 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr11_-_25324534 | 0.22 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr10_-_43718914 | 0.22 |
ENSDART00000189277
|
cetn3
|
centrin 3 |
| chr17_+_38476300 | 0.22 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr14_-_24110707 | 0.22 |
ENSDART00000133522
ENSDART00000123152 |
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr3_-_26787430 | 0.21 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr14_-_2209742 | 0.20 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr23_-_18707418 | 0.20 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr17_-_7028418 | 0.20 |
ENSDART00000188305
ENSDART00000187895 |
sash1b
|
SAM and SH3 domain containing 1b |
| chr4_-_64284924 | 0.20 |
ENSDART00000166733
|
CT956002.2
|
|
| chr11_-_37880492 | 0.19 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr9_+_6587364 | 0.19 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr5_+_37785152 | 0.19 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr3_-_53533128 | 0.18 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr1_-_40123943 | 0.18 |
ENSDART00000146917
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr13_-_13754091 | 0.18 |
ENSDART00000131255
|
ky
|
kyphoscoliosis peptidase |
| chr24_+_10310577 | 0.18 |
ENSDART00000141718
|
otulina
|
OTU deubiquitinase with linear linkage specificity a |
| chr18_+_22217489 | 0.17 |
ENSDART00000165464
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr11_+_24046179 | 0.17 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr7_+_31879649 | 0.17 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr3_+_45758760 | 0.17 |
ENSDART00000076007
|
gper1
|
G protein-coupled estrogen receptor 1 |
| chr6_-_42971184 | 0.16 |
ENSDART00000014552
|
arl8ba
|
ADP-ribosylation factor-like 8Ba |
| chr8_+_17884569 | 0.16 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr7_-_33881275 | 0.16 |
ENSDART00000100102
|
rxfp3.3a1
|
relaxin/insulin-like family peptide receptor 3.3a1 |
| chr14_-_26425416 | 0.16 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr20_-_31427390 | 0.16 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr8_-_40251126 | 0.15 |
ENSDART00000180435
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr11_+_16152316 | 0.15 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr9_+_30720048 | 0.15 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr23_-_31506854 | 0.14 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr4_-_13255700 | 0.14 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr12_-_43435254 | 0.13 |
ENSDART00000182723
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr3_+_53156813 | 0.13 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr10_-_7386475 | 0.13 |
ENSDART00000167963
|
nrg1
|
neuregulin 1 |
| chr6_-_39765546 | 0.12 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr12_+_25945560 | 0.12 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr24_+_25919809 | 0.12 |
ENSDART00000006615
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr3_+_15776446 | 0.12 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
| chr1_+_23784905 | 0.12 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr3_+_17456428 | 0.11 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr10_-_34889053 | 0.11 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr13_+_30874153 | 0.11 |
ENSDART00000112380
ENSDART00000189016 ENSDART00000134809 |
ercc6
|
excision repair cross-complementation group 6 |
| chr19_-_35596207 | 0.11 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr23_+_42346799 | 0.10 |
ENSDART00000159985
ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr14_+_5385855 | 0.10 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr10_+_8534750 | 0.10 |
ENSDART00000183960
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr13_+_11829072 | 0.10 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr20_+_54336137 | 0.09 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr18_+_5213338 | 0.08 |
ENSDART00000033574
|
slc24a5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr4_-_72609735 | 0.08 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr9_-_19699728 | 0.08 |
ENSDART00000166780
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr8_-_14067517 | 0.07 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr3_-_50118140 | 0.07 |
ENSDART00000131913
|
hgs
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr20_+_15982482 | 0.07 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr7_-_38687431 | 0.07 |
ENSDART00000074490
|
aplnr2
|
apelin receptor 2 |
| chr14_-_10387377 | 0.07 |
ENSDART00000145118
|
commd5
|
COMM domain containing 5 |
| chr3_+_23703704 | 0.07 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr1_+_54835131 | 0.05 |
ENSDART00000145070
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr4_-_12104421 | 0.05 |
ENSDART00000139561
|
mrps33
|
mitochondrial ribosomal protein S33 |
| chr25_+_20694177 | 0.04 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr15_-_23475051 | 0.04 |
ENSDART00000152460
|
nlrx1
|
NLR family member X1 |
| chr8_+_18010568 | 0.04 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr15_-_4023182 | 0.04 |
ENSDART00000190195
|
CR626884.5
|
|
| chr23_-_25779995 | 0.04 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr15_-_8763106 | 0.03 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr1_+_16574312 | 0.03 |
ENSDART00000187067
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr24_-_17400143 | 0.03 |
ENSDART00000134947
|
cul1b
|
cullin 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf5a+atf5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.3 | 0.8 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.3 | 0.8 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.3 | 0.8 | GO:0035992 | tendon formation(GO:0035992) |
| 0.2 | 1.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.2 | 0.9 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.2 | 1.1 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.2 | 0.5 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 0.9 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 0.5 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 1.5 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.7 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 1.6 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.4 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.5 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 2.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.1 | 0.7 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.4 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 0.7 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.2 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.1 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.5 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.4 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.1 | GO:0036135 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 1.1 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 1.2 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 1.0 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.1 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 1.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.5 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 1.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.1 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.9 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.1 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 2.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 3.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 0.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 2.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.7 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.9 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 1.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.2 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |