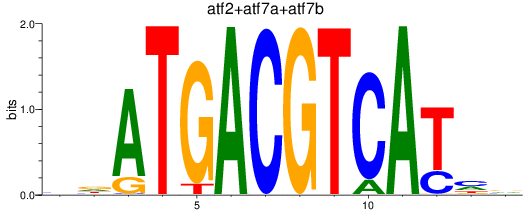
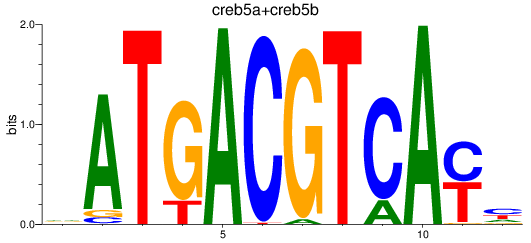
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for atf2+atf7a+atf7b_creb5a+creb5b
Z-value: 2.65
Transcription factors associated with atf2+atf7a+atf7b_creb5a+creb5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf7a
|
ENSDARG00000011298 | activating transcription factor 7a |
|
atf2
|
ENSDARG00000023903 | activating transcription factor 2 |
|
atf7b
|
ENSDARG00000055481 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000114492 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000115171 | activating transcription factor 7b |
|
creb5b
|
ENSDARG00000070536 | cAMP responsive element binding protein 5b |
|
creb5a
|
ENSDARG00000099002 | cAMP responsive element binding protein 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf2 | dr11_v1_chr9_+_2343096_2343172 | 0.95 | 1.3e-02 | Click! |
| atf7b | dr11_v1_chr6_-_39631164_39631181 | 0.85 | 6.8e-02 | Click! |
| creb5b | dr11_v1_chr16_-_20707742_20707742 | 0.74 | 1.5e-01 | Click! |
| creb5a | dr11_v1_chr19_-_19505167_19505167 | 0.55 | 3.3e-01 | Click! |
| atf7a | dr11_v1_chr23_-_27479558_27479558 | 0.19 | 7.6e-01 | Click! |
Activity profile of atf2+atf7a+atf7b_creb5a+creb5b motif
Sorted Z-values of atf2+atf7a+atf7b_creb5a+creb5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43606502 | 4.50 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr3_-_49566364 | 3.83 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr9_+_7548533 | 3.55 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr24_-_7632187 | 3.50 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr16_+_34523515 | 3.42 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr6_+_13920479 | 3.32 |
ENSDART00000155480
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr8_+_44714336 | 3.11 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr1_-_22861348 | 2.82 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr9_-_296169 | 2.80 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr19_-_1961024 | 2.71 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr16_-_12173554 | 2.63 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr7_+_40228422 | 2.56 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr13_-_50672341 | 2.51 |
ENSDART00000190498
|
FP102122.1
|
|
| chr6_-_48311 | 2.51 |
ENSDART00000131010
|
zgc:114175
|
zgc:114175 |
| chr25_+_35683956 | 2.48 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr13_+_36146415 | 2.42 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr17_+_8184649 | 2.39 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr1_-_38756870 | 2.36 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr2_+_35603637 | 2.28 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr6_+_27667359 | 2.28 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr7_-_12065668 | 2.26 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr20_+_29743904 | 2.22 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr1_+_41666611 | 2.19 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr10_-_24371312 | 2.17 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr23_+_23183449 | 2.16 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr17_+_29345606 | 2.15 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr21_-_23308286 | 2.15 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr2_+_20332044 | 2.14 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr24_-_24163201 | 2.12 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr15_+_28685625 | 2.12 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr11_-_44163164 | 2.11 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr8_+_29267093 | 2.09 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr15_+_28685892 | 2.06 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr2_-_42415902 | 2.03 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr16_-_32649929 | 2.00 |
ENSDART00000136161
|
faxcb
|
failed axon connections homolog b |
| chr3_-_2593859 | 1.99 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr20_+_20637866 | 1.97 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr23_-_29667716 | 1.93 |
ENSDART00000158302
ENSDART00000133902 |
clstn1
|
calsyntenin 1 |
| chr13_-_40499296 | 1.91 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr3_-_5964557 | 1.89 |
ENSDART00000184738
|
BX284638.1
|
|
| chr24_-_24162930 | 1.87 |
ENSDART00000080602
|
map7d2b
|
MAP7 domain containing 2b |
| chr20_+_20638034 | 1.86 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr5_-_23999777 | 1.82 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr8_+_29742237 | 1.81 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr13_+_23843712 | 1.75 |
ENSDART00000057611
|
oprm1
|
opioid receptor, mu 1 |
| chr2_+_20331445 | 1.74 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr16_+_5184402 | 1.71 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr17_-_26911852 | 1.70 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr8_+_21588067 | 1.67 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr23_-_29667544 | 1.65 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr19_+_2279051 | 1.65 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr21_-_23307653 | 1.61 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr2_-_56635744 | 1.59 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr20_+_25911342 | 1.55 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr15_+_5028608 | 1.46 |
ENSDART00000092809
|
abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr9_+_219124 | 1.45 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr8_-_9118958 | 1.43 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr13_+_32446169 | 1.41 |
ENSDART00000143325
|
nt5c1ba
|
5'-nucleotidase, cytosolic IB a |
| chr13_+_28705143 | 1.41 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr1_+_10305611 | 1.40 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr15_-_11341635 | 1.40 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr14_+_790166 | 1.40 |
ENSDART00000123912
|
adra2da
|
adrenergic, alpha-2D-, receptor a |
| chr23_-_4915118 | 1.39 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr21_-_3770636 | 1.38 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr13_-_40754499 | 1.35 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr10_+_44903676 | 1.29 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr16_-_9869056 | 1.29 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr10_+_37145007 | 1.23 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr16_+_25137483 | 1.20 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr17_+_23554932 | 1.19 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr15_+_1004680 | 1.18 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr4_-_69189894 | 1.18 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr20_-_34801181 | 1.17 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr14_+_597532 | 1.17 |
ENSDART00000159805
|
LO018568.2
|
|
| chr20_-_18915376 | 1.16 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr18_+_8917766 | 1.16 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr2_+_38147761 | 1.15 |
ENSDART00000135307
|
sall2
|
spalt-like transcription factor 2 |
| chr18_-_19103929 | 1.15 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr6_-_48473395 | 1.15 |
ENSDART00000185096
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr9_-_13963078 | 1.14 |
ENSDART00000193398
ENSDART00000061156 |
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr5_-_30145939 | 1.13 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr9_-_306515 | 1.12 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr7_+_7552008 | 1.10 |
ENSDART00000173018
ENSDART00000049311 |
clcn3
|
chloride channel 3 |
| chr12_-_5455936 | 1.09 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr20_+_9211237 | 1.09 |
ENSDART00000139527
|
si:ch211-59d15.4
|
si:ch211-59d15.4 |
| chr12_-_32066469 | 1.09 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr1_-_17693273 | 1.08 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr2_-_30324610 | 1.05 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr18_+_12058403 | 1.04 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr17_-_15382704 | 1.03 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr25_+_28893615 | 1.02 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr22_+_2239254 | 1.02 |
ENSDART00000131396
ENSDART00000135320 |
znf1144
|
zinc finger protein 1144 |
| chr24_-_19718077 | 1.00 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr18_+_910992 | 0.99 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr14_+_11950011 | 0.98 |
ENSDART00000188138
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr21_-_25395223 | 0.98 |
ENSDART00000016219
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr14_-_48939560 | 0.97 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr2_+_22694382 | 0.97 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr5_-_41494831 | 0.95 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr3_+_20156956 | 0.95 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr6_-_39893501 | 0.94 |
ENSDART00000141611
ENSDART00000135631 ENSDART00000077662 ENSDART00000130613 |
myl6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr12_-_30443562 | 0.93 |
ENSDART00000020769
|
adrb1
|
adrenoceptor beta 1 |
| chr25_-_7686201 | 0.92 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr7_+_24889783 | 0.92 |
ENSDART00000005329
ENSDART00000159955 |
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr15_-_12319065 | 0.91 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr15_-_20412286 | 0.90 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr18_+_17663898 | 0.90 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr21_-_32374656 | 0.89 |
ENSDART00000112550
|
mapk9
|
mitogen-activated protein kinase 9 |
| chr7_-_12909352 | 0.89 |
ENSDART00000172901
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr20_-_24122881 | 0.88 |
ENSDART00000131857
|
bach2b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
| chr12_+_7491690 | 0.88 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr12_-_35505610 | 0.87 |
ENSDART00000105518
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr24_+_4373033 | 0.87 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr24_+_4373355 | 0.87 |
ENSDART00000179062
ENSDART00000093256 ENSDART00000138943 |
ccny
|
cyclin Y |
| chr17_+_16873417 | 0.87 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr16_-_33097398 | 0.86 |
ENSDART00000166617
|
dopey1
|
dopey family member 1 |
| chr8_-_25846188 | 0.86 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr7_-_39203799 | 0.86 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr15_+_618376 | 0.86 |
ENSDART00000156007
|
si:ch73-144d13.8
|
si:ch73-144d13.8 |
| chr8_+_39607466 | 0.86 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr7_+_30493684 | 0.83 |
ENSDART00000027466
|
mindy2
|
MINDY lysine 48 deubiquitinase 2 |
| chr5_+_57743815 | 0.81 |
ENSDART00000005090
|
alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr10_+_6496185 | 0.79 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr16_-_41131578 | 0.79 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr7_+_23907692 | 0.79 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr21_-_5007109 | 0.78 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr24_-_41657005 | 0.78 |
ENSDART00000159109
|
CABZ01044099.1
|
|
| chr13_-_45475289 | 0.78 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr11_-_22372072 | 0.77 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr23_-_17509656 | 0.77 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr18_-_7143920 | 0.77 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr5_-_24000211 | 0.76 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr20_-_32446406 | 0.76 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr18_-_46063773 | 0.73 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr15_-_27710513 | 0.73 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr24_-_18876877 | 0.73 |
ENSDART00000186269
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr15_-_591308 | 0.73 |
ENSDART00000153884
|
si:ch73-144d13.5
|
si:ch73-144d13.5 |
| chr24_+_744713 | 0.73 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr17_-_20979077 | 0.72 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr13_-_31470439 | 0.71 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr15_-_28200049 | 0.71 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr17_-_8674208 | 0.71 |
ENSDART00000149201
|
ctbp2a
|
C-terminal binding protein 2a |
| chr1_-_54947592 | 0.70 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr11_+_28166165 | 0.70 |
ENSDART00000169360
ENSDART00000192311 |
ephb2b
|
eph receptor B2b |
| chr19_+_42227400 | 0.69 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr16_+_46111849 | 0.69 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr24_-_18877118 | 0.68 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr16_-_16212615 | 0.67 |
ENSDART00000059905
|
upp1
|
uridine phosphorylase 1 |
| chr8_-_410199 | 0.67 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr19_-_6873107 | 0.66 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr21_+_9576176 | 0.66 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr15_-_163586 | 0.63 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr21_-_275377 | 0.63 |
ENSDART00000157509
|
rln1
|
relaxin 1 |
| chr3_-_1204341 | 0.63 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr23_-_25686894 | 0.63 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr4_+_76403698 | 0.62 |
ENSDART00000184821
ENSDART00000169373 |
FP074874.2
FP074874.1
|
|
| chr25_-_12906872 | 0.62 |
ENSDART00000165156
ENSDART00000167449 |
sept15
|
septin 15 |
| chr14_+_16813816 | 0.60 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr4_+_40953320 | 0.60 |
ENSDART00000151912
|
znf1136
|
zinc finger protein 1136 |
| chr1_+_9290103 | 0.60 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr18_-_12451772 | 0.60 |
ENSDART00000175083
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr4_+_359970 | 0.60 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr1_-_11291324 | 0.59 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr6_+_21227621 | 0.59 |
ENSDART00000193583
|
prkca
|
protein kinase C, alpha |
| chr21_+_45366229 | 0.59 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr25_+_7321675 | 0.57 |
ENSDART00000104712
ENSDART00000142934 |
hmg20a
|
high mobility group 20A |
| chr22_+_38935060 | 0.57 |
ENSDART00000183732
ENSDART00000130055 |
sirt7
|
sirtuin 7 |
| chr4_-_73825089 | 0.57 |
ENSDART00000174207
|
si:dkey-262g12.12
|
si:dkey-262g12.12 |
| chr4_-_16124417 | 0.55 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr23_-_27505825 | 0.55 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr6_-_25952848 | 0.55 |
ENSDART00000076997
ENSDART00000148748 |
lmo4b
|
LIM domain only 4b |
| chr5_+_26921033 | 0.55 |
ENSDART00000051483
|
tm2d2
|
TM2 domain containing 2 |
| chr5_+_69808763 | 0.54 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr1_+_49878000 | 0.54 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr5_-_32882162 | 0.53 |
ENSDART00000085769
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr5_-_32363372 | 0.53 |
ENSDART00000098045
|
gas1b
|
growth arrest-specific 1b |
| chr8_-_24113575 | 0.52 |
ENSDART00000099692
ENSDART00000186211 |
dclre1b
|
DNA cross-link repair 1B |
| chr13_+_15816573 | 0.52 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr25_-_31763897 | 0.51 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr15_-_34845414 | 0.51 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr2_+_42200390 | 0.51 |
ENSDART00000012919
|
ift57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr7_-_27686021 | 0.50 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr6_+_58832155 | 0.50 |
ENSDART00000144842
|
dctn2
|
dynactin 2 (p50) |
| chr20_+_41549200 | 0.50 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr4_+_33373100 | 0.49 |
ENSDART00000150417
|
znf1090
|
zinc finger protein 1090 |
| chr15_-_923981 | 0.49 |
ENSDART00000155285
|
si:dkey-77f5.14
|
si:dkey-77f5.14 |
| chr9_-_28939181 | 0.49 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_-_30324297 | 0.49 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr22_+_30330574 | 0.48 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr22_+_2801464 | 0.48 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr7_-_28568238 | 0.48 |
ENSDART00000173927
|
tmem9b
|
TMEM9 domain family, member B |
| chr15_+_46386261 | 0.48 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr8_-_39984593 | 0.48 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr12_+_48204891 | 0.47 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr17_+_12698532 | 0.47 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr13_-_3155243 | 0.46 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr4_-_73739119 | 0.45 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr4_-_30362840 | 0.45 |
ENSDART00000165929
|
znf1083
|
zinc finger protein 1083 |
| chr8_-_13678415 | 0.45 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr4_-_30422325 | 0.45 |
ENSDART00000158444
|
znf1114
|
zinc finger protein 1114 |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf2+atf7a+atf7b_creb5a+creb5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 2.8 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.6 | 2.3 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.5 | 2.7 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.4 | 2.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 1.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 1.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.4 | 1.5 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.3 | 4.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 3.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.3 | 1.4 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 0.7 | GO:0090190 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 0.7 | GO:0072526 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.9 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 1.0 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.2 | 2.3 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.4 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 1.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.9 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.9 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 2.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.6 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.5 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 5.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 2.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 1.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.5 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.4 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 2.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.7 | GO:0044206 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 1.4 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.7 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.2 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.1 | 0.6 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 4.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 3.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.4 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.2 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.2 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 11.2 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0006670 | sphingosine metabolic process(GO:0006670) diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 1.6 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.4 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.2 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.9 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 3.2 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.2 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 2.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.4 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 2.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.8 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 1.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.4 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 2.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.2 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 4.5 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 1.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 4.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 2.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 2.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 0.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 7.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 6.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 9.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 3.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 8.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.5 | 4.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 2.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 1.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.9 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.2 | 3.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 0.8 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.5 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.8 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 1.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 2.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 1.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.6 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 3.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 0.9 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 4.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.5 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 2.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 1.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 2.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 10.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.2 | GO:0015562 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 1.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.4 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 4.1 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 2.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |